Transient Complexity of E. coli Lipidome Is Explained by Fatty Acyl Synthesis and Cyclopropanation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Culture Growth and Growth Phase Identification of E. coli Strains
2.2. Microscopy of E. coli Cells
2.3. The Total Lipid and Total Protein Extraction
2.4. Identification and Quantification of Lipids
2.5. HILIC-MS2 Data Processing
2.6. GC-MS Data Processing
2.7. CFA Synthase Western Blot Assay
3. Results
3.1. Growth Phase as a Basis for Lipidomic Comparison between Strains
3.2. Lipidome Dynamics Is Determined by Synthesis and Cyclopropanation
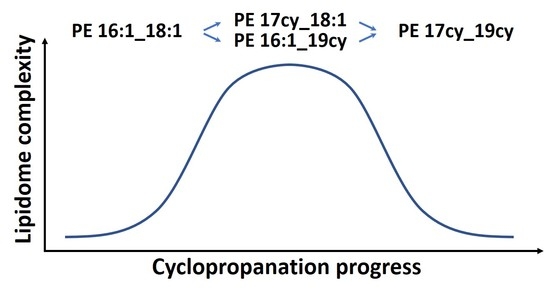
3.3. Cyclopropanation Dynamics of Lipids with Unsaturated Chains and Transient Diversity
3.4. Dynamics of the Saturated to Unsaturated Lipid Ratio—A Surrogate for Fluidity
3.5. Membrane Surface Charge Dynamics
3.6. E. coli Envelope Dynamics
3.7. Effects of Antibiotics
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Blattner, F.R.; Plunkett, G.; Bloch, C.A.; Perna, N.T.; Burland, V.; Riley, M.; Collado-Vides, J.; Glasner, J.D.; Rode, C.K.; Mayhew, G.F.; et al. The Complete Genome Sequence of Escherichia coli K-12. Science 1997, 277, 1453–1462. [Google Scholar] [CrossRef]
- Jeucken, A.; Molenaar, M.R.; van de Lest, C.H.A.; Jansen, J.W.A.; Helms, J.B.; Brouwers, J.F. A Comprehensive Functional Characterization of Escherichia coli Lipid Genes. Cell Rep. 2019, 27, 1597–1606.e2. [Google Scholar] [CrossRef]
- Ishihama, Y.; Schmidt, T.; Rappsilber, J.; Mann, M.; Hartl, F.U.; Kerner, M.J.; Frishman, D. Protein abundance profiling of the Escherichia coli cytosol. BMC Genom. 2008, 9, 102. [Google Scholar] [CrossRef]
- Taniguchi, Y.; Choi, P.J.; Li, G.-W.; Chen, H.; Babu, M.; Hearn, J.; Emili, A.; Xie, X.S. Quantifying E. coli Proteome and Transcriptome with Single-Molecule Sensitivity in Single Cells. Science 2010, 329, 533–538. [Google Scholar] [CrossRef]
- Oursel, D.; Loutelier-Bourhis, C.; Orange, N.; Chevalier, S.; Norris, V.; Lange, C.M. Lipid composition of membranes of Escherichia coli by liquid chromatography/tandem mass spectrometry using negative electrospray ionization. Rapid Commun. Mass Spectrom. 2007, 21, 1721–1728. [Google Scholar] [CrossRef]
- Oursel, D.; Loutelier-Bourhis, C.; Orange, N.; Chevalier, S.; Norris, V.; Lange, C.M. Identification and relative quantification of fatty acids in Escherichia coli membranes by gas chromatography/mass spectrometry. Rapid Commun. Mass Spectrom. 2007, 21, 3229–3233. [Google Scholar] [CrossRef]
- Dowhan, W. A retrospective: Use of Escherichia coli as a vehicle to study phospholipid synthesis and function. Biochim. Biophys. Acta (BBA)—Mol. Cell Biol. Lipids 2013, 1831, 471–494. [Google Scholar] [CrossRef]
- Jeucken, A.; Zhou, M.; Wösten, M.M.S.M.; Brouwers, J.F. Control of n-Butanol Induced Lipidome Adaptations in E. coli. Metabolites 2021, 11, 286. [Google Scholar] [CrossRef]
- Kralj, T.; Nuske, M.; Hofferek, V.; Sani, M.-A.; Lee, T.-H.; Separovic, F.; Aguilar, M.-I.; Reid, G.E. Multi-Omic Analysis to Characterize Metabolic Adaptation of the E. coli Lipidome in Response to Environmental Stress. Metabolites 2022, 12, 171. [Google Scholar] [CrossRef]
- Rowlett, V.W.; Mallampalli, V.K.P.S.; Karlstaedt, A.; Dowhan, W.; Taegtmeyer, H.; Margolin, W.; Vitrac, H. Impact of Membrane Phospholipid Alterations in Escherichia coli on Cellular Function and Bacterial Stress Adaptation. J. Bacteriol. 2017, 199, e00849-00816. [Google Scholar] [CrossRef] [Green Version]
- Wehrli, P.M.; Angerer, T.B.; Farewell, A.; Fletcher, J.S.; Gottfries, J. Investigating the Role of the Stringent Response in Lipid Modifications during the Stationary Phase in E. coli by Direct Analysis with Time-of-Flight-Secondary Ion Mass Spectrometry. Anal. Chem. 2016, 88, 8680–8688. [Google Scholar] [CrossRef] [PubMed]
- Oh, H.Y.; Lee, J.O.; Kim, O.B. Increase of organic solvent tolerance of Escherichia coli by the deletion of two regulator genes, fadR and marR. Appl. Microbiol. Biotechnol. 2012, 96, 1619–1627. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.-Y.; Cronan, J.E. Membrane cyclopropane fatty acid content is a major factor in acid resistance of Escherichia coli. Mol. Microbiol. 1999, 33, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Raetz, C.R. Enzymology, genetics, and regulation of membrane phospholipid synthesis in Escherichia coli. Microbiol. Rev. 1978, 42, 614–659. [Google Scholar] [CrossRef] [PubMed]
- Raetz, C.R.; Dowhan, W. Biosynthesis and function of phospholipids in Escherichia coli. J. Biol. Chem. 1990, 265, 1235–1238. [Google Scholar] [CrossRef]
- Hines, K.M.; Xu, L. Lipidomic consequences of phospholipid synthesis defects in Escherichia coli revealed by HILIC-ion mobility-mass spectrometry. Chem. Phys. Lipids 2019, 219, 15–22. [Google Scholar] [CrossRef]
- Erridge, C.; Bennett-Guerrero, E.; Poxton, I.R. Structure and function of lipopolysaccharides. Microbes Infect. 2002, 4, 837–851. [Google Scholar] [CrossRef]
- Sohlenkamp, C.; Geiger, O. Bacterial membrane lipids: Diversity in structures and pathways. FEMS Microbiol. Rev. 2016, 40, 133–159. [Google Scholar] [CrossRef]
- Parsons, J.B.; Rock, C.O. Bacterial lipids: Metabolism and membrane homeostasis. Prog. Lipid Res. 2013, 52, 249–276. [Google Scholar] [CrossRef]
- Cronan, J.E.; Thomas, J. Chapter 17 Bacterial Fatty Acid Synthesis and its Relationships with Polyketide Synthetic Pathways. In Methods in Enzymology; Academic Press: New York, NY, USA, 2009; Volume 459, pp. 395–433. [Google Scholar]
- Jeucken, A.; Helms, J.B.; Brouwers, J.F. Cardiolipin synthases of Escherichia coli have phospholipid class specific phospholipase D activity dependent on endogenous and foreign phospholipids. Biochim. Biophys. Acta Mol. Cell. Biol. Lipids 2018, 1863, 1345–1353. [Google Scholar] [CrossRef]
- Li, C.; Tan, B.K.; Zhao, J.; Guan, Z. In Vivo and in Vitro Synthesis of Phosphatidylglycerol by an Escherichia coli Cardiolipin Synthase. J. Biol. Chem. 2016, 291, 25144–25153. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Lin, Y.; Lu, S.; Zhang, J.; Bogdanov, M. Biogenesis, transport and remodeling of lysophospholipids in Gram-negative bacteria. Biochim. Biophys. Acta Mol. Cell. Biol. Lipids 2017, 1862, 1404–1413. [Google Scholar] [CrossRef] [PubMed]
- Grogan, D.W.; Cronan, J.E. Cyclopropane ring formation in membrane lipids of bacteria. Microbiol. Mol. Biol. Rev. 1997, 61, 429–441. [Google Scholar] [PubMed]
- Wang, A.Y.; Cronan, J.E., Jr. The growth phase-dependent synthesis of cyclopropane fatty acids in Escherichia coli is the result of an RpoS(KatF)-dependent promoter plus enzyme instability. Mol. Microbiol. 1994, 11, 1009–1017. [Google Scholar] [CrossRef] [PubMed]
- Hari, S.B.; Grant, R.A.; Sauer, R.T. Structural and Functional Analysis of E. coli Cyclopropane Fatty Acid Synthase. Structure 2018, 26, 1251–1258.e53. [Google Scholar] [CrossRef] [PubMed]
- Poger, D.; Mark, A.E. A Ring to Rule Them All: The Effect of Cyclopropane Fatty Acids on the Fluidity of Lipid Bilayers. J. Phys. Chem. B 2015, 119, 5487–5495. [Google Scholar] [CrossRef] [PubMed]
- Fontecave, M.; Atta, M.; Mulliez, E. S-adenosylmethionine: Nothing goes to waste. Trends Biochem. Sci. 2004, 29, 243–249. [Google Scholar] [CrossRef]
- Liebisch, G.; Fahy, E.; Aoki, J.; Dennis, E.A.; Durand, T.; Ejsing, C.S.; Fedorova, M.; Feussner, I.; Griffiths, W.J.; Köfeler, H.; et al. Update on LIPID MAPS classification, nomenclature, and shorthand notation for MS-derived lipid structures. J. Lipid Res. 2020, 61, 1539–1555. [Google Scholar] [CrossRef]
- Chang, Y.-Y.; Eichel, J.; Cronan, J.E. Metabolic Instability of Escherichia coli Cyclopropane Fatty Acid Synthase Is Due to RpoH-Dependent Proteolysis. J. Bacteriol. 2000, 182, 4288–4294. [Google Scholar] [CrossRef]
- Hildebrand, J.G.; Law, J.H. Fatty Acid Distribution in Bacterial Phospholipids. The Specificity of the Cyclopropane Synthetase Reaction*. Biochemistry 1964, 3, 1304–1308. [Google Scholar] [CrossRef]
- Cronan, J.E. Phospholipid Alterations During Growth of Escherichia coli. J. Bacteriol. 1968, 95, 2054–2061. [Google Scholar] [CrossRef] [PubMed]
- Shibuya, I.; Yamagoe, S.; Miyazaki, C.; Matsuzaki, H.; Ohta, A. Biosynthesis of novel acidic phospholipid analogs in Escherichia coli. J. Bacteriol. 1985, 161, 473–477. [Google Scholar] [CrossRef] [PubMed]
- Raetz, C.R.H. Molecular genetics of membrane phospholipid synthesis. Annu. Rev. Genet. 1986, 20, 253–291. [Google Scholar] [CrossRef]
- Garrett, T.A.; Raetz, C.R.; Richardson, T.; Kordestani, R.; Son, J.D.; Rose, R.L. Identification of phosphatidylserylglutamate: A novel minor lipid in Escherichia coli. J. Lipid Res. 2009, 50, 1589–1599. [Google Scholar] [CrossRef] [PubMed]
- Gidden, J.; Denson, J.; Liyanage, R.; Ivey, D.M.; Lay, J.O. Lipid compositions in Escherichia coli and Bacillus subtilis during growth as determined by MALDI-TOF and TOF/TOF mass spectrometry. Int. J. Mass Spectrom. 2009, 283, 178–184. [Google Scholar] [CrossRef] [PubMed]
- Garrett, T.A.; Raetz, C.R.H.; Son, J.D.; Richardson, T.D.; Bartling, C.; Guan, Z. Non-enzymatically derived minor lipids found in Escherichia coli lipid extracts. Biochim. Biophys. Acta (BBA)—Mol. Cell Biol. Lipids 2011, 1811, 827–837. [Google Scholar] [CrossRef]
- Blevins, M.S.; Klein, D.R.; Brodbelt, J.S. Localization of Cyclopropane Modifications in Bacterial Lipids via 213 nm Ultraviolet Photodissociation Mass Spectrometry. Anal. Chem. 2019, 91, 6820–6828. [Google Scholar] [CrossRef]
- Chung, H.J.; Bang, W.; Drake, M.A. Stress Response of Escherichia coli. Compr. Rev. Food Sci. Food Saf. 2006, 5, 52–64. [Google Scholar] [CrossRef]
- Epand, R.F.; Savage, P.B.; Epand, R.M. Bacterial lipid composition and the antimicrobial efficacy of cationic steroid compounds (Ceragenins). Biochim. Biophys. Acta (BBA)—Biomembr. 2007, 1768, 2500–2509. [Google Scholar] [CrossRef]
- Alves, E.; Santos, N.; Melo, T.; Maciel, E.; Dória, M.L.; Faustino, M.A.F.; Tomé, J.P.C.; Neves, M.G.P.M.S.; Cavaleiro, J.A.S.; Cunha, Â.; et al. Photodynamic oxidation of Escherichia coli membrane phospholipids: New insights based on lipidomics. Rapid Commun. Mass Spectrom. 2013, 27, 2717–2728. [Google Scholar] [CrossRef]
- Calvano, C.D.; Picca, R.A.; Bonerba, E.; Tantillo, G.; Cioffi, N.; Palmisano, F. MALDI-TOF mass spectrometry analysis of proteins and lipids in Escherichia coli exposed to copper ions and nanoparticles. J. Mass Spectrom. 2016, 51, 828–840. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Gonzalez, L.E.; Ferreira, C.R.; Vorsilak, A.; Frabutt, D.; Sobreira, T.J.P.; Pugia, M.; Cooks, R.G. Multiple Reaction Monitoring Profiling (MRM-Profiling) of Lipids To Distinguish Strain-Level Differences in Microbial Resistance in Escherichia coli. Anal. Chem. 2019, 91, 11349–11354. [Google Scholar] [CrossRef] [PubMed]
- Vaidyanathan, S.; Kell, D.B.; Goodacre, R. Flow-injection electrospray ionization mass spectrometry of crude cell extracts for high-throughput bacterial identification. J. Am. Soc. Mass Spectrom. 2002, 13, 118–128. [Google Scholar] [CrossRef]
- Oliver, P.M.; Crooks, J.A.; Leidl, M.; Yoon, E.J.; Saghatelian, A.; Weibel, D.B. Localization of Anionic Phospholipids in Escherichia coli Cells. J. Bacteriol. 2014, 196, 3386–3398. [Google Scholar] [CrossRef]
- Bogdanov, M.; Pyrshev, K.; Yesylevskyy, S.; Ryabichko, S.; Boiko, V.; Ivanchenko, P.; Kiyamova, R.; Guan, Z.; Ramseyer, C.; Dowhan, W. Phospholipid distribution in the cytoplasmic membrane of Gram-negative bacteria is highly asymmetric, dynamic, and cell shape-dependent. Sci. Adv. 2020, 6, eaaz6333. [Google Scholar] [CrossRef]
- Wang, Q.; Ding, Y.; Liu, L.; Shi, J.; Sun, J.; Xue, Y. Engineering Escherichia coli for autoinducible production of n-butanol. Electron. J. Biotechnol. 2015, 18, 138–142. [Google Scholar] [CrossRef]
- Chilambi, G.S.; Hinks, J.; Matysik, A.; Zhu, X.; Choo, P.Y.; Liu, X.; Chan-Park, M.B.; Bazan, G.C.; Kline, K.A.; Rice, S.A. Enterococcus faecalis Adapts to Antimicrobial Conjugated Oligoelectrolytes by Lipid Rearrangement and Differential Expression of Membrane Stress Response Genes. Front. Microbiol. 2020, 11, 155. [Google Scholar] [CrossRef]
- Zhou, C.; Chia, G.W.N.; Ho, J.C.S.; Moreland, A.S.; Seviour, T.; Liedberg, B.; Parikh, A.N.; Kjelleberg, S.; Hinks, J.; Bazan, G.C. A Chain-Elongated Oligophenylenevinylene Electrolyte Increases Microbial Membrane Stability. Adv. Mater. 2019, 31, 1808021. [Google Scholar] [CrossRef]
- Limwongyut, J.; Liu, Y.; Chilambi, G.S.; Seviour, T.; Hinks, J.; Mu, Y.; Bazan, G.C. Interactions of a paracyclophane-based conjugated oligoelectrolyte with biological membranes. RSC Adv. 2018, 8, 39849–39853. [Google Scholar] [CrossRef]
- Hinks, J.; Wang, Y.; Matysik, A.; Kraut, R.; Kjelleberg, S.; Mu, Y.; Bazan, G.C.; Wuertz, S.; Seviour, T. Increased Microbial Butanol Tolerance by Exogenous Membrane Insertion Molecules. ChemSusChem 2015, 8, 3718–3726. [Google Scholar] [CrossRef]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef] [PubMed]
- Samantha, A.; Vrielink, A. Lipid A Phosphoethanolamine Transferase: Regulation, Structure and Immune Response. J. Mol. Biol. 2020, 432, 5184–5196. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Ji, S.; Burla, B.; Wenk, M.R.; Torta, F.; Cazenave-Gassiot, A. LICAR: An Application for Isotopic Correction of Targeted Lipidomic Data Acquired with Class-Based Chromatographic Separations Using Multiple Reaction Monitoring. Anal. Chem. 2021, 93, 3163–3171. [Google Scholar] [CrossRef]
- Dunn, W.B.; Broadhurst, D.; Begley, P.; Zelena, E.; Francis-McIntyre, S.; Anderson, N.; Brown, M.; Knowles, J.D.; Halsall, A.; Haselden, J.N.; et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2011, 6, 1060–1083. [Google Scholar] [CrossRef] [PubMed]
- Budin, I.; de Rond, T.; Chen, Y.; Chan, L.J.G.; Petzold, C.J.; Keasling, J.D. Viscous control of cellular respiration by membrane lipid composition. Science 2018, 362, 1186–1189. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-M.; Rock, C.O. Membrane lipid homeostasis in bacteria. Nat. Rev. Microbiol. 2008, 6, 222–233. [Google Scholar] [CrossRef] [PubMed]
- Morein, S.; Andersson, A.-S.; Rilfors, L.; Lindblom, G. Wild-type Escherichia coli Cells Regulate the Membrane Lipid Composition in a Window between Gel and Non-lamellar Structures. J. Biol. Chem. 1996, 271, 6801–6809. [Google Scholar] [CrossRef]
- Bishop, R.E.; Gibbons, H.S.; Guina, T.; Trent, M.S.; Miller, S.I.; Raetz, C.R.H. Transfer of palmitate from phospholipids to lipid A in outer membranes of Gram-negative bacteria. EMBO J. 2000, 19, 5071–5080. [Google Scholar] [CrossRef]
- Keller, R.; Ariöz, C.; Hansmeier, N.; Stenberg-Bruzell, F.; Burstedt, M.; Vikström, D.; Kelly, A.; Wieslander, Å.; Daley, D.O.; Hunke, S. The Escherichia coli Envelope Stress Sensor CpxA Responds to Changes in Lipid Bilayer Properties. Biochemistry 2015, 54, 3670–3676. [Google Scholar] [CrossRef]
- Anaya-López, J.L.; López-Meza, J.E.; Ochoa-Zarzosa, A. Bacterial resistance to cationic antimicrobial peptides. Crit. Rev. Microbiol. 2013, 39, 180–195. [Google Scholar] [CrossRef]
- Raetz, C.R.H.; Reynolds, C.M.; Trent, M.S.; Bishop, R.E. Lipid A modification systems in gram-negative bacteria. Annu. Rev. Biochem. 2007, 76, 295–329. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Chia, G.W.N.; Berezhnoy, N.V.; Cazenave-Gassiot, A.; Kjelleberg, S.; Hinks, J.; Mu, Y.; Seviour, T. Bacterial lipopolysaccharide core structures mediate effects of butanol ingress. Biochim. Biophys. Acta (BBA)—Biomembr. 2020, 1862, 183150. [Google Scholar] [CrossRef] [PubMed]
- Klein, S.; Lorenzo, C.; Hoffmann, S.; Walther, J.M.; Storbeck, S.; Piekarski, T.; Tindall, B.J.; Wray, V.; Nimtz, M.; Moser, J. Adaptation of Pseudomonas aeruginosa to various conditions includes tRNA-dependent formation of alanyl-phosphatidylglycerol. Mol. Microbiol. 2009, 71, 551–565. [Google Scholar] [CrossRef] [PubMed]
- Volkmer, B.; Heinemann, M. Condition-Dependent Cell Volume and Concentration of Escherichia coli to Facilitate Data Conversion for Systems Biology Modeling. PLoS ONE 2011, 6, e23126. [Google Scholar] [CrossRef] [PubMed]
- Jun, S.; Si, F.; Pugatch, R.; Scott, M. Fundamental principles in bacterial physiology—History, recent progress, and the future with focus on cell size control: A review. Rep. Prog. Phys. 2018, 81, 056601. [Google Scholar] [CrossRef]
- Hinks, J.; Poh, W.H.; Chu, J.J.H.; Loo, J.S.C.; Bazan, G.C.; Hancock, L.E.; Wuertz, S. Oligopolyphenylenevinylene-Conjugated Oligoelectrolyte Membrane Insertion Molecules Selectively Disrupt Cell Envelopes of Gram-Positive Bacteria. Appl. Environ. Microbiol. 2015, 81, 1949–1958. [Google Scholar] [CrossRef]
- Simpson, R.B.; Ashbrook, J.D.; Santos, E.C.; Spector, A.A. Partition of fatty acids. J. Lipid Res. 1974, 15, 415–422. [Google Scholar] [CrossRef]
- Kim, B.H.; Kim, S.; Kim, H.G.; Lee, J.; Lee, I.S.; Park, Y.K. The formation of cyclopropane fatty acids in Salmonella enterica serovar Typhimurium. Microbiology 2005, 151, 209–218. [Google Scholar] [CrossRef]
- Machida, S.; Shiraiwa, Y.; Suzuki, I. Construction of a cyanobacterium synthesizing cyclopropane fatty acids. Biochim. Biophys. Acta (BBA)—Mol. Cell Biol. Lipids 2016, 1861, 980–987. [Google Scholar] [CrossRef]
- Nyström, T. Stationary-Phase Physiology. Annu. Rev. Microbiol. 2004, 58, 161–181. [Google Scholar] [CrossRef]
- Navarro Llorens, J.M.; Tormo, A.; Martínez-García, E. Stationary phase in gram-negative bacteria. FEMS Microbiol. Rev. 2010, 34, 476–495. [Google Scholar] [CrossRef] [PubMed]
- Makarova, M.; Peter, M.; Balogh, G.; Glatz, A.; MacRae, J.I.; Lopez Mora, N.; Booth, P.; Makeyev, E.; Vigh, L.; Oliferenko, S. Delineating the Rules for Structural Adaptation of Membrane-Associated Proteins to Evolutionary Changes in Membrane Lipidome. Curr. Biol. 2020, 30, 367–380.e368. [Google Scholar] [CrossRef] [PubMed]
- Szenk, M.; Dill, K.A.; de Graff, A.M.R. Why Do Fast-Growing Bacteria Enter Overflow Metabolism? Testing the Membrane Real Estate Hypothesis. Cell Syst. 2017, 5, 95–104. [Google Scholar] [CrossRef]
- Davis, B.D.; Luger, S.M.; Tai, P.C. Role of ribosome degradation in the death of starved Escherichia coli cells. J. Bacteriol. 1986, 166, 439–445. [Google Scholar] [CrossRef] [PubMed]
- Vadia, S.; Tse, J.L.; Lucena, R.; Yang, Z.; Kellogg, D.R.; Wang, J.D.; Levin, P.A. Fatty Acid Availability Sets Cell Envelope Capacity and Dictates Microbial Cell Size. Curr. Biol. 2017, 27, 1757–1767.e1755. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, Q.; Guan, S.; Shi, H. Quantitative Connection between Cell Size and Growth Rate by Phospholipid Metabolism. Cells 2020, 9, 391. [Google Scholar] [CrossRef] [Green Version]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Berezhnoy, N.V.; Cazenave-Gassiot, A.; Gao, L.; Foo, J.C.; Ji, S.; Regina, V.R.; Yap, P.K.P.; Wenk, M.R.; Kjelleberg, S.; Seviour, T.W.; et al. Transient Complexity of E. coli Lipidome Is Explained by Fatty Acyl Synthesis and Cyclopropanation. Metabolites 2022, 12, 784. https://0-doi-org.brum.beds.ac.uk/10.3390/metabo12090784
Berezhnoy NV, Cazenave-Gassiot A, Gao L, Foo JC, Ji S, Regina VR, Yap PKP, Wenk MR, Kjelleberg S, Seviour TW, et al. Transient Complexity of E. coli Lipidome Is Explained by Fatty Acyl Synthesis and Cyclopropanation. Metabolites. 2022; 12(9):784. https://0-doi-org.brum.beds.ac.uk/10.3390/metabo12090784
Chicago/Turabian StyleBerezhnoy, Nikolay V., Amaury Cazenave-Gassiot, Liang Gao, Juat Chin Foo, Shanshan Ji, Viduthalai Rasheedkhan Regina, Pui Khee Peggy Yap, Markus R. Wenk, Staffan Kjelleberg, Thomas William Seviour, and et al. 2022. "Transient Complexity of E. coli Lipidome Is Explained by Fatty Acyl Synthesis and Cyclopropanation" Metabolites 12, no. 9: 784. https://0-doi-org.brum.beds.ac.uk/10.3390/metabo12090784