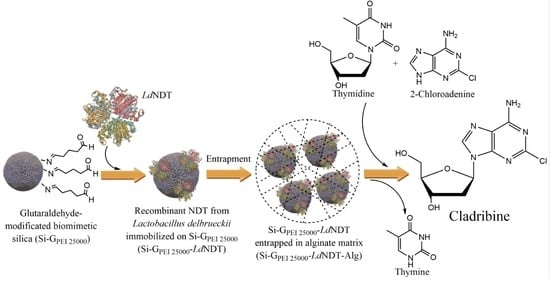
Green Production of Cladribine by Using Immobilized 2′-Deoxyribosyltransferase from Lactobacillus delbrueckii Stabilized through a Double Covalent/Entrapment Technology
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Production of LdNDT
2.3. Standard Activity Assay for LdNDT
2.4. LdNDT Immobilization
2.4.1. Biomimetic Silica Entrapment (SiBio)
2.4.2. Immobilization on Modified Biomimetic Silica Nanoparticles
2.4.3. Cyanogen Bromide (CNBr) Immobilization
2.5. Biochemical Characterization of SiGPEI25000-LdNDT Derivative
2.6. Derivative Entrapment in Alginate
2.7. Surface Morphology Study
2.8. Thermal Inactivation and pH Stability
2.9. Storage Stability and Operational Reusability
2.10. Bioprocess Scale-Up
2.11. Analytical Methods
2.12. Molecular Modeling
2.13. Sustainability Impact
3. Results and Discussion
3.1. Enzyme Immobilization Screening
3.2. Synthesis of SiG Nanoparticles Using Several PEI Sizes
3.3. Optimization of Reaction Parameters
3.4. SiGPEI25000-LdNDT Stabilization by Calcium Alginate Entrapment
3.4.1. Temperature and pH Stability
3.4.2. Scanning Electron Microscopy (SEM) Analysis
3.5. Storage Stability and Reusability of SiGPEI25000-LdNDT and SiGPEI25000-LdNDT-Alg Biocatalysts
3.6. Bioprocess Scale-Up
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Spurgeon, S.; Yu, M.; Phillips, J.D.; Epner, E.M. Cladribine: Not just another purine analogue? Expert Opin. Investig. Drugs 2002, 18, 1169–1181. [Google Scholar] [CrossRef] [PubMed]
- Biernacki, T.; Sandi, D.; Bencsik, K.; Vécsei, L. Medicinal chemistry of multiple sclerosis: Focus on cladribine. Mini Rev. Med. Chem. 2020, 20, 269–285. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y. A practical synthesis of 2-chloro-2’-deoxyadenosine (cladribine) from 2’-deoxyadenosine. J. Chem. Res. 2013, 37, 213–215. [Google Scholar] [CrossRef]
- Fernández-Lucas, J.; Camarasa, M.J. (Eds.) Enzymatic and Chemical Synthesis of Nucleic Acid Derivatives; John Wiley & Sons: Hoboken, NJ, USA, 2019. [Google Scholar] [CrossRef]
- Lapponi, M.J.; Rivero, C.W.; Zinni, M.A.; Britos, C.N.; Trelles, J.A. New developments in nucleoside analogues biosynthesis: A review. J. Mol. Catal. B Enzym. 2016, 133, 218–233. [Google Scholar] [CrossRef]
- Fresco-Taboada, A.; de La Mata, I.; Arroyo, M.; Fernández-Lucas, J. New insights on nucleoside 2’-deoxyribosyltransferases: A versatile biocatalyst for one-pot one-step synthesis of nucleoside analogs. Appl. Microbiol. Biotechnol. 2013, 97, 3773–3785. [Google Scholar] [CrossRef]
- Del Arco, J.; Acosta, J.; Fernández-Lucas, J. New trends in the biocatalytic production of nucleosidic active pharmaceutical ingredients using 2’-deoxyribosyltransferases. Biotechnol. Adv. 2021, 107701. [Google Scholar] [CrossRef]
- Mikhailopulo, I.A.; Miroshnikov, A.I. Biologically important nucleosides: Modern trends in biotechnology and application. Mendeleev Commun. 2011, 21, 57–69. [Google Scholar] [CrossRef]
- Yehia, H.; Kamel, S.; Paulick, K.; Neubauer, P.; Wagner, A. Substrate spectra of nucleoside phosphorylases and their potential in the production of pharmaceutically active compounds. Curr. Pharm. Des. 2017, 23, 6913–6935. [Google Scholar] [CrossRef]
- Méndez, M.B.; Rivero, C.W.; López-Gallego, F.; Guisán, J.M.; Trelles, J.A. Development of a high efficient biocatalyst by oriented covalent immobilization of a novel recombinant 2’-N-deoxyribosyltransferase from Lactobacillus animalis. J. Biotechnol. 2018, 270, 39–43. [Google Scholar] [CrossRef]
- Britos, C.N.; Lapponi, M.J.; Cappa, V.A.; Rivero, C.W.; Trelles, J.A. Biotransformation of halogenated nucleosides by immobilized Lactobacillus animalis 2′-N-deoxyribosyltransferase. J. Fluor. Chem. 2016, 186, 91–96. [Google Scholar] [CrossRef]
- Drenichev, M.S.; Alexeev, C.S.; Kurochkin, N.N.; Mikhailov, S.N. Use of nucleoside phosphorylases for the preparation of purine and pyrimidine 2’-deoxynucleosides. Adv. Synth. Catal. 2018, 360, 305–312. [Google Scholar] [CrossRef]
- Liu, G.; Cheng, T.; Chu, J.; Li, S.; He, B. Efficient synthesis of purine nucleoside analogs by a new trimeric purine nucleoside phosphorylase from Aneurinibacillusmigulanus AM007. Molecules 2020, 25, 100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rabuffetti, M.; Bavaro, T.; Semproli, R.; Cattaneo, G.; Massone, M.; Morelli, C.F.; Speranza, G.; Ubiali, D. Synthesis of ribavirin, tecadenoson, and cladribine by enzymatic transglycosylation. Catalysts 2019, 9, 355. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.; Szeker, K.; Jiao, L.Y.; Oestreich, M.; Mikhailopulo, I.A.; Neubauer, P. Synthesis of 2,6-dihalogenated purine nucleosides by thermostable nucleoside phosphorylases. Adv. Synth. Catal. 2015, 357, 1237–1244. [Google Scholar] [CrossRef]
- Sheldon, R.A.; van Pelt, S. Enzyme immobilization in biocatalysis: Why, what and how. Chem. Soc. Rev. 2013, 42, 6223–6235. [Google Scholar] [CrossRef] [Green Version]
- Mateo, C.; Palomo, J.M.; Fernandez-Lorente, G.; Guisan, J.M.; Fernández-Lafuente, R. Improvement of enzyme activity, stability and selectivity via immobilization techniques. Enzyme Microb. Technol. 2007, 40, 1451–1463. [Google Scholar] [CrossRef]
- Barbosa, O.; Torres, R.; Ortiz, C.; Berenguer-Murcia, A.; Rodrigues, R.C.; Fernandez-Lafuente, R. Heterofunctional supports in enzyme immobilization: From traditional immobilization protocols to opportunities in tuning enzyme properties. Biomacromolecules 2013, 14, 2433–2462. [Google Scholar] [CrossRef] [Green Version]
- Rodrigues, R.C.; Ortiz, C.; Berenguer-Murcia, Á.; Torres, R.; Fernández-Lafuente, R. Modifying enzyme activity and selectivity by immobilization. Chem. Soc. Rev. 2013, 42, 6290–6307. [Google Scholar] [CrossRef]
- Guisan, J.M.; Lopez-Gallego, F.; Bolivar, J.M.; Rocha-Martín, J.; Fernandez-Lorente, G. The science of enzyme immobilization. In Immobilization of Enzymes and Cells; Humana Press: Totowa, NJ, USA, 2020; pp. 1–26. [Google Scholar] [CrossRef]
- Rimola, A.; Costa, D.; Sodupe, M.; Lambert, J.F.; Ugliengo, P. Silica surface features and their role in the adsorption of biomolecules: Computational modeling and experiments. Chem. Rev. 2013, 113, 4216–4313. [Google Scholar] [CrossRef] [Green Version]
- Zucca, P.; Sanjust, E. Inorganic materials as supports for covalent enzyme immobilization: Methods and mechanisms. Molecules 2014, 19, 14139–14194. [Google Scholar] [CrossRef]
- Luckarift, H.R.; Spain, J.C.; Naik, R.R.; Stone, M.O. Enzyme immobilization in a biomimetic silica support. Nat. Biotechnol. 2004, 22, 211–213. [Google Scholar] [CrossRef] [PubMed]
- Coradin, T.; Lopez, P.J.; Gautier, C.; Livage, J. From biogenic to biomimetic silica. C. R. Palevol 2004, 3, 443–452. [Google Scholar] [CrossRef]
- Sumper, M.; Brunner, E. Silica biomineralisation in diatoms: The model organism Thalassiosirapseudonana. ChemBioChem 2008, 9, 1187–1194. [Google Scholar] [CrossRef]
- Lechner, C.C.; Becker, C.F.W. Silaffins in silica biomineralization and biomimetic silica precipitation. Mar. Drugs 2015, 13, 5297–5333. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.Y.; Mooney, D.J. Alginate: Properties, biomedical applications. Prog. Polym. Sci. 2012, 37, 106–126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Funami, T.; Fang, T.; Noda, S.; Ishihara, S.; Nakauma, M.; Draget, K.I.; Nishinari, K.; Phillips, G.O. Rheological properties of sodium alginate in an aqueous system during gelation in relation to supermolecular structures and Ca2+ binding. Food Hydrocoll. 2009, 23, 1746–1755. [Google Scholar] [CrossRef]
- Cappa, V.A.; Rivero, C.W.; Britos, C.N.; Martinez, L.M.; Lozano, M.E.; Trelles, J.A. An efficient biocatalytic system for floxuridine biosynthesis based on Lactobacillus animalis ATCC 35046 immobilized in Sr-alginate. Process.Biochem. 2014, 49, 1169–1175. [Google Scholar] [CrossRef]
- Acosta, J.; del Arco, J.; Martinez-Pascual, S.; Clemente-Suárez, V.J.; Fernández-Lucas, J. One-pot multi-enzymatic production of purine derivatives with application in pharmaceutical and food industry. Catalysts 2018, 8, 9. [Google Scholar] [CrossRef] [Green Version]
- Acosta, J.; Pérez, E.; Sánchez-Murcia, P.A.; Fillat, C.; Fernández-Lucas, J. Molecular basis of NDT-mediated activation of nucleoside-based prodrugs and application in suicide gene therapy. Biomolecules 2021, 11, 120. [Google Scholar] [CrossRef]
- Cazaban, D.; Illanes, A.; Wilson, L.; Betancor, L. Bio-inspired silica lipase nanobiocatalysts for the synthesis of fatty acid methyl esters. Process Biochem. 2018, 74, 86–93. [Google Scholar] [CrossRef]
- Trelles, J.A.; Rivero, C.W. Whole cell entrapment techniques. In Immobilization of Enzymes and Cells; Humana Press: Totowa, NJ, USA, 2020; pp. 385–394. [Google Scholar] [CrossRef]
- Delano, W.L. The PyMOL Molecular Graphics System; DeLano Scientific: Palo Alto, CA, USA, 2002. [Google Scholar]
- Acosta, J.; del Arco, J.; Pisabarro, V.; Gago, F.; Fernández-Lucas, J. N-ribosyltransferase from Archaeoglobusveneficus: A novel halotolerant and thermostable biocatalyst for the synthesis of purine ribonucleoside analogs. Front. Bioeng. Biotechnol. 2020, 8, 593. [Google Scholar] [CrossRef] [PubMed]
- Acosta, J.; del Arco, J.; del Pozo, M.L.; Herrera-Tapias, B.; Clemente-Suárez, V.J.; Berenguer, J.; Hidalgo, A.; Fernández-Lucas, J. Hypoxanthine-guanine phosphoribosyltransferase/adenylate kinase from Zobelliagalactanivorans: A bifunctional catalyst for the synthesis of nucleoside-5-mono-, di-and triphosphates. Front. Bioeng. Biotechnol. 2020, 8, 677. [Google Scholar] [CrossRef] [PubMed]
- Roe, D.R.; Cheatham, T.E., III. PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef] [PubMed]
- Rivero, C.W.; de Benedetti, E.C.; Sambeth, J.; Trelles, J.A. Biotransformation of cladribine by a nanostabilized extremophilic biocatalyst. J. Biotechnol. 2020, 323, 166–173. [Google Scholar] [CrossRef] [PubMed]
- Zucca, P.; Fernandez-Lafuente, R.; Sanjust, E. Agarose and its derivatives as supports for enzyme immobilization. Molecules 2016, 21, 1577. [Google Scholar] [CrossRef]
- Betancor, L.; Luckarift, H.R. Bioinspired enzyme encapsulation for biocatalysis. Trends Biotechnol. 2008, 26, 566–572. [Google Scholar] [CrossRef] [PubMed]
- Jackson, E.; Correa, S.; Betancor, L. In situ immobilization of enzymes in biomimetic silica. In Immobilization of Enzymes and Cells; Humana Press: Totowa, NJ, USA, 2020; pp. 259–270. [Google Scholar] [CrossRef]
- Fresco-Taboada, A.; Serra, I.; Fernández-Lucas, J.; Acebal, C.; Arroyo, M.; Terreni, M.; de la Mata, I. Nucleoside 2’-deoxyribosyltransferase from psychrophilic bacterium Bacillus psychrosaccharolyticus—Preparation of an immobilized biocatalyst for the enzymatic synthesis of therapeutic nucleosides. Molecules 2014, 19, 11231–11249. [Google Scholar] [CrossRef] [Green Version]
- Fresco-Taboada, A.; Serra, I.; Arroyo, M.; Fernández-Lucas, J.; de la Mata, I.; Terreni, M. Development of an immobilized biocatalyst based on Bacillus psychrosaccharolyticus NDT for the preparative synthesis of trifluridine and decytabine. Catal. Today 2016, 259, 197–204. [Google Scholar] [CrossRef]
- Barbosa, O.; Ortiz, C.; Berenguer-Murcia, Á.; Torres, R.; Rodrigues, R.C.; Fernandez-Lafuente, R. Glutaraldehyde in biocatalysts design: A useful crosslinker and a versatile tool in enzyme immobilization. RSC Adv. 2014, 4, 1583–1600. [Google Scholar] [CrossRef] [Green Version]
- Pérez, E.; Sánchez-Murcia, P.A.; Jordaan, J.; Blanco, M.D.; Mancheño, J.M.; Gago, F.; Fernández-Lucas, J. Enzymatic synthesis of therapeutic nucleosides using a highly versatile purine nucleoside 2’-deoxyribosyltransferase from Trypanosoma brucei. ChemCatChem 2018, 10, 4406–4416. [Google Scholar] [CrossRef]
- Fernández-Lucas, J.; Harris, R.; Mata-Casar, I.; Heras, A.; de la Mata, I.; Arroyo, M. Magnetic chitosan beads for covalent immobilization of nucleoside 2’-deoxyribosyltransferase: Application in nucleoside analogues synthesis. J. Ind. Microbiol. Biotechnol. 2013, 40, 955–966. [Google Scholar] [CrossRef]
- Virgen-Ortíz, J.J.; Dos Santos, J.C.; Berenguer-Murcia, Á.; Barbosa, O.; Rodrigues, R.C.; Fernandez-Lafuente, R. Polyethylenimine: A very useful ionic polymer in the design of immobilized enzyme biocatalysts. J. Mater. Chem. B 2017, 5, 7461–7490. [Google Scholar] [CrossRef] [Green Version]
- Rivero, C.W.; de Benedetti, E.C.; López Gallego, F.; Pessela, B.C.; Guisán, J.M.; Trelles, J.A. Biosynthesis of an antiviral compound using a stabilized phosphopentomutase by multipoint covalent immobilization. J. Biotechnol. 2017, 249, 34–41. [Google Scholar] [CrossRef]
- Del Arco, J.; Pérez, E.; Naitow, H.; Matsuura, Y.; Kunishima, N.; Fernández-Lucas, J. Structural and functional characterization of thermostable biocatalysts for the synthesis of 6-aminopurine nucleoside-5’-monophospate analogues. Bioresour. Technol. 2019, 276, 244–252. [Google Scholar] [CrossRef]
- Del Arco, J.; Martínez-Pascual, S.; Clemente-Suárez, V.J.; Corral, O.J.; Jordaan, J.; Hormigo, D.; Perona, A.; Fernández-Lucas, J. One-pot, one-step production of dietary nucleotides by magnetic biocatalysts. Catalysts 2018, 8, 184. [Google Scholar] [CrossRef] [Green Version]
- Del Arco, J.; Galindo, J.; Clemente-Suárez, V.J.; Corrales, A.; Fernández-Lucas, J. Sustainable synthesis of uridine-5’-monophosphate analogues by immobilized uracil phosphoribosyltransferase from Thermus thermophilus. Biochim. Biophys. Acta 2020, 1868, 140251. [Google Scholar] [CrossRef] [PubMed]
- Notley, S.M.; Leong, Y.K. Interaction between silica in the presence of adsorbed poly(ethyleneimine): Correlation between colloidal probe adhesion measurements and yield stress. Phys. Chem. Chem. Phys. 2010, 12, 10594–10601. [Google Scholar] [CrossRef]
- Iliescu, R.I.; Andronescu, E.; Voicu, G.; Ficai, A.; Covaliu, C.I. Hybrid materials based on montmorillonite and citostatic drugs: Preparation and characterization. Appl. Clay Sci. 2011, 52, 62–68. [Google Scholar] [CrossRef]
- Lapponi, M.J.; Britos, C.N.; Rivero, C.W.; Trelles, J.A. Biotransformation of cladribine using a stabilized biocatalyst in calcium alginate beads. Biotechnol. Progr. 2020, 36, e2927. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Lucas, J.; Condezo, L.A.; Quezada, M.A.; Sinisterra, J.V. Low-temperature synthesis of 2’-deoxyadenosine using immobilized psychrotrophic microorganisms. Biotechnol. Bioengineer. 2008, 100, 213–222. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Lucas, J.; Condezo, L.A.; Martinez-Lagos, F.; Sinisterra, J.V.; Biotransformation Group. Synthesis of 2’-deoxyibosylnucleosides using new 2’-deoxyribosyltransferase microorganism producers. Enzym. Microbial. Technol. 2007, 40, 1147–1155. [Google Scholar] [CrossRef]
- Crespo, N.; Sánchez-Murcia, P.A.; Gago, F.; Cejudo-Sanches, J.; Galmes, M.A.; Fernández-Lucas, J.; Mancheño, J.M. 2’-Deoxyribosyltransferase from Leishmania mexicana, an efficient biocatalyst for one-pot, one-step synthesis of nucleosides from poorly soluble purine bases. Appl. Microbiol. Biotechnol. 2017, 101, 7187–7200. [Google Scholar] [CrossRef] [Green Version]
- Del Arco, J.; Cejudo-Sanches, J.; Esteban, I.; Clemente-Suárez, V.J.; Hormigo, D.; Perona, A.; Fernández-Lucas, J. Enzymatic production of dietary nucleotides from low-soluble purine bases by an efficient, thermostable and alkali-tolerant biocatalyst. Food Chem. 2017, 237, 605–611. [Google Scholar] [CrossRef]
- Jackson, E.; Ferrari, M.; Cuestas-Ayllon, C.; Fernández-Pacheco, R.; Perez-Carvajal, J.; de la Fuente, J.M.; Grazú, V.; Betancor, L. Protein-templated biomimetic silica nanoparticles. Langmuir 2015, 31, 3687–3695. [Google Scholar] [CrossRef]
- Rinaldi, F.; Fernández-Lucas, J.; de la Fuente, D.; Zheng, C.; Bavaro, T.; Peters, B.; Massolini, G.; Annunziata, F.; Conti, P.; de la Mata, I.; et al. Immobilized enzyme reactors based on nucleoside phosphorylases and 2’-deoxyribosyltransferase for the in-flow synthesis of pharmaceutically relevant nucleoside ana-logues. Bioresour. Technol. 2020, 307, 123258. [Google Scholar] [CrossRef]
- Del Arco, J.; Jordaan, J.; Moral-Dardé, V.; Fernández-Lucas, J. Sustainable production of nucleoside analogues by a high-efficient purine 2’-deoxyribosyltransferase immobilized onto Ni2+ chelate magnetic microparticles. Bioresour. Technol. 2019, 289, 121772. [Google Scholar] [CrossRef] [PubMed]
- Taran, S.A.; Verevkina, K.N.; Feofanov, S.A.; Miroshnikov, A.I. Enzymatic transglycosylation of natural and modified nucleosides by immobilized thermostable nucleoside phosphorylases from Geobacillus stearothermophilus. Russ. J. Bioorganic Chem. 2009, 35, 739–745. [Google Scholar] [CrossRef] [PubMed]
- De Benedetti, E.C.; Rivero, C.W.; Britos, C.N.; Lozano, M.E.; Trelles, J.A. Biotransformation of 2,6-diaminopurine nucleosides by immobilized Geobacillus stearothermophilus. Biotechnol. Progr. 2012, 28, 1251–1256. [Google Scholar] [CrossRef] [PubMed]
- Sheldon, R.A. Green chemistry and resource efficiency: Towards a green economy. Green Chem. 2016, 18, 3180–3183. [Google Scholar] [CrossRef]







| Biocatalyst | Immobilization (%) | Specific Activity (IU/mg) a | Retained Activity (%) |
|---|---|---|---|
| LdNDT | |||
| 230 µg b | 70 | 2.9 ± 0.1 | 94 |
| SiGPEI1200-1300-LdNDT | 3.0 ± 0.1 | 97 | |
| 15 µg b | 100 | ||
| 95 µg b | 97 | ||
| 230 µg b | 83 | ||
| 470 µg b | 50 | ||
| SiGPEI25000-LdNDT | 3.0 ± 0.1 | 97 | |
| 15 µg b | 99 | ||
| 95 µg b | 97 | ||
| 230 µg b | 88 | ||
| 470 µg b | 55 | ||
| SiGPEI70000-LdNDT | 3.0 ± 0.1 | 97 | |
| 15 µg b | 98 | ||
| 95 µg b | 97 | ||
| 230 µg b | 81 | ||
| 470 µg b | 50 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rivero, C.W.; García, N.S.; Fernández-Lucas, J.; Betancor, L.; Romanelli, G.P.; Trelles, J.A. Green Production of Cladribine by Using Immobilized 2′-Deoxyribosyltransferase from Lactobacillus delbrueckii Stabilized through a Double Covalent/Entrapment Technology. Biomolecules 2021, 11, 657. https://0-doi-org.brum.beds.ac.uk/10.3390/biom11050657
Rivero CW, García NS, Fernández-Lucas J, Betancor L, Romanelli GP, Trelles JA. Green Production of Cladribine by Using Immobilized 2′-Deoxyribosyltransferase from Lactobacillus delbrueckii Stabilized through a Double Covalent/Entrapment Technology. Biomolecules. 2021; 11(5):657. https://0-doi-org.brum.beds.ac.uk/10.3390/biom11050657
Chicago/Turabian StyleRivero, Cintia Wanda, Natalia Soledad García, Jesús Fernández-Lucas, Lorena Betancor, Gustavo Pablo Romanelli, and Jorge Abel Trelles. 2021. "Green Production of Cladribine by Using Immobilized 2′-Deoxyribosyltransferase from Lactobacillus delbrueckii Stabilized through a Double Covalent/Entrapment Technology" Biomolecules 11, no. 5: 657. https://0-doi-org.brum.beds.ac.uk/10.3390/biom11050657