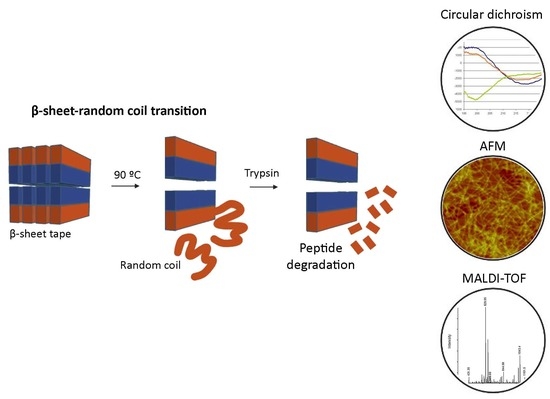
β-Sheet to Random Coil Transition in Self-Assembling Peptide Scaffolds Promotes Proteolytic Degradation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Circular Dichroism (CD)
2.2. Atomic Force Microscopy (AFM)
2.3. MALDI-TOF
3. Results
3.1. Structural Characterization of the Self-Assembling Peptide RAD16-I
3.2. Enzymatic Degradation of RAD16-I Assessed by MALDI-TOF
3.3. Enzymatic Degradation of RAD16-I Assessed by AFM
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Moffat, K.L.; Neal, R.A.; Freed, L.E.; Guilak, F. Engineering Functional Tissues: In Vitro Culture Parameters. In Principles of Tissue Engineering, 4th ed.; Lanza, R., Langer, R., Vacanti, J., Eds.; Elsevier: Amsterdam, The Netherlands, 2013; pp. 237–259. [Google Scholar] [CrossRef]
- Kim, S.-K.; Jang, S.D.; Kim, H.; Chung, S.; Park, J.K.; Kuh, H.-J. Phenotypic Heterogeneity and Plasticity of Cancer Cell Migration in a Pancreatic Tumor Three-Dimensional Culture Model. Cancers 2020, 12, 1305. [Google Scholar] [CrossRef] [PubMed]
- Griffith, L.G.; Swartz, M.A. Capturing complex 3D tissue physiology in vitro. Nat. Rev. Mol. Cell Biol. 2006, 7, 211–224. [Google Scholar] [CrossRef] [PubMed]
- Ratheesh, G.; Venugopal, J.R.; Chinappan, A.; Ezhilarasu, H.; Sadiq, A.; Ramakrishna, S. 3D Fabrication of Polymeric Scaffolds for Regenerative Therapy. ACS Biomater. Sci. Eng. 2017, 3, 1175–1194. [Google Scholar] [CrossRef] [PubMed]
- Nagase, H.; Visse, R.; Murphy, G. Structure and function of matrix metalloproteinases and TIMPs. Cardiovasc. Res. 2006, 69, 562–573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonnans, C.; Chou, J.; Werb, Z. Remodelling the extracellular matrix in development and disease. Nat. Rev. Mol. Cell Biol. 2014, 15, 786–801. [Google Scholar] [CrossRef]
- Elsawy, M.A.; Kim, K.-H.; Park, J.-W.; Deep, A. Hydrolytic degradation of polylactic acid (PLA) and its composites. Renew. Sustain. Energy Rev. 2017, 79, 1346–1352. [Google Scholar] [CrossRef]
- Samantaray, P.K.; Little, A.; Haddleton, D.M.; McNally, T.; Tan, B.; Sun, Z.; Huang, W.; Ji, Y.; Wan, C. Poly(glycolic acid) (PGA): A versatile building block expanding high performance and sustainable bioplastic applications. Green Chem. 2020, 22, 4055–4081. [Google Scholar] [CrossRef]
- Erdal, N.B.; Lando, G.A.; Yadav, A.; Srivastava, R.K.; Hakkarainen, M. Hydrolytic Degradation of Porous Crosslinked Poly(ε-Caprolactone) Synthesized by High Internal Phase Emulsion Templating. Polymers 2020, 12, 1849. [Google Scholar] [CrossRef] [PubMed]
- Le Fer, G.; Becker, M.L. 4D Printing of Resorbable Complex Shape-Memory Poly(propylene fumarate) Star Scaffolds. ACS Appl. Mater. Interfaces 2020, 12, 22444–22452. [Google Scholar] [CrossRef]
- Sung, H.J.; Meredith, C.; Johnson, C.; Galis, Z.S. The effect of scaffold degradation rate on three-dimensional cell growth and angiogenesis. Biomaterials 2004, 25, 5735–5742. [Google Scholar] [CrossRef]
- Wade, R.J.; Bassin, E.J.; Rodell, C.B.; Burdick, J.A. Protease-degradable electrospun fibrous hydrogels. Nat. Commun. 2015, 6, 6639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.; Zhang, Y.; Hou, Z.; Cui, X.; Zhao, Y.; Xu, H. Rational Design of Short Peptide-Based Hydrogels with MMP-2 Responsiveness for Controlled Anticancer Peptide Delivery. Biomacromolecules 2017, 18, 3563–3571. [Google Scholar] [CrossRef] [PubMed]
- He, Y.J.; Young, D.A.; Mededovic, M.; Li, K.; Li, C.; Tichauer, K.; Venerus, D.; Papavasiliou, G. Protease-Sensitive Hydrogel Biomaterials with Tunable Modulus and Adhesion Ligand Gradients for 3D Vascular Sprouting. Biomacromolecules 2018, 19, 4168–4181. [Google Scholar] [CrossRef] [PubMed]
- González-Pérez, F.; Ibáñez-Fonseca, A.; Alonso, M.; Rodríguez-Cabello, J.C. Combining tunable proteolytic sequences and a VEGF-mimetic peptide for the spatiotemporal control of angiogenesis within Elastin-Like Recombinamer scaffolds. Acta Biomater. 2021, 130, 149–160. [Google Scholar] [CrossRef] [PubMed]
- Beamish, J.A.; Juliar, B.A.; Cleveland, D.S.; Busch, M.E.; Nimmagadda, L.; Putnam, A.J. Deciphering the relative roles of matrix metalloproteinase- and plasmin-mediated matrix degradation during capillary morphogenesis using engineered hydrogels. J. Biomed. Mater. Res. Part B Appl. Biomater. 2019, 107, 2507–2516. [Google Scholar] [CrossRef] [PubMed]
- Coletta, D.J.; Ibáñez-Fonseca, A.; Missana, L.R.; Jammal, M.V.; Vitelli, E.J.; Aimone, M.; Zabalza, F.; Issa, J.P.M.; Alonso, M.; Rodríguez-Cabello, J.C.; et al. Bone Regeneration Mediated by a Bioactive and Biodegradable Extracellular Matrix-Like Hydrogel Based on Elastin-Like Recombinamers. Tissue Eng. Part A 2017, 23, 1361–1371. [Google Scholar] [CrossRef] [PubMed]
- Szilágyi, B.Á.; Némethy, Á.; Magyar, A.; Szabó, I.; Bősze, S.; Gyarmati, B.; Szilágyi, A. Amino acid based polymer hydrogel with enzymatically degradable cross-links. React. Funct. Polym. 2018, 133, 21–28. [Google Scholar] [CrossRef]
- Kim, K.; Bae, B.; Kang, Y.J.; Nam, J.-M.; Kang, S.; Ryu, J.-H. Natural Polypeptide-Based Supramolecular Nanogels for Stable Noncovalent Encapsulation. Biomacromolecules 2013, 14, 3515–3522. [Google Scholar] [CrossRef]
- Yao, Q.; Kou, L.; Tu, Y.; Zhu, L. MMP-Responsive ‘Smart’ Drug Delivery and Tumor Targeting. Trends Pharmacol. Sci. 2018, 39, 766–781. [Google Scholar] [CrossRef] [PubMed]
- Betriu, N.; Recha-Sancho, L.; Semino, C.E. Culturing Mammalian Cells in Three-dimensional Peptide Scaffolds. J. Vis. Exp. 2018, e57259. [Google Scholar] [CrossRef]
- Semino, C.E. Self-assembling Peptides: From Bio-inspired Materials to Bone Regeneration. J. Dent. Res. 2008, 87, 606–616. [Google Scholar] [CrossRef] [PubMed]
- Sieminski, A.L.; Semino, C.E.; Gong, H.; Kamm, R.D. Primary sequence of ionic self-assembling peptide gels affects endothelial cell adhesion and capillary morphogenesis. J. Biomed. Mater. Res. Part A 2008, 87, 494–504. [Google Scholar] [CrossRef]
- Genové, E.; Schmitmeier, S.; Sala, A.; Borrós, S.; Bader, A.; Griffith, L.G.; Semino, C.E. Functionalized self-assembling peptide hydrogel enhance maintenance of hepatocyte activity in vitro. J. Cell Mol. Med. 2009, 13, 3387–3397. [Google Scholar] [CrossRef] [Green Version]
- Fernández-Muiños, T.; Recha-Sancho, L.; López-Chicón, P.; Castells-Sala, C.; Mata, A.; Semino, C.E. Bimolecular based heparin and self-assembling hydrogel for tissue engineering applications. Acta Biomater. 2015, 16, 35–48. [Google Scholar] [CrossRef] [PubMed]
- Betriu, N.; Semino, C.E. Development of a 3D Co-Culture System as a Cancer Model Using a Self-Assembling Peptide Scaffold. Gels 2018, 4, 65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Betriu, N.; Andreeva, A.; Semino, C.E. Erlotinib Promotes Ligand-Induced EGFR Degradation in 3D but Not 2D Cultures of Pancreatic Ductal Adenocarcinoma Cells. Cancers 2021, 13, 4504. [Google Scholar] [CrossRef] [PubMed]
- Recha-Sancho, L.; Semino, C.E. Chondroitin sulfate- and decorin-based self-Assembling scaffolds for cartilage tissue engineering. PLoS ONE 2016, 11, e0157603. [Google Scholar] [CrossRef]
- Betriu, N.; Jarrosson-Moral, C.; Semino, C.E. Culture and Differentiation of Human Hair Follicle Dermal Papilla Cells in a Soft 3D Self-Assembling Peptide Scaffold. Biomolecules 2020, 10, 684. [Google Scholar] [CrossRef]
- Castells-Sala, C.; Recha-Sancho, L.; Llucià-Valldeperas, A.; Soler-Botija, C.; Bayes-Genis, A.; Semino, C.E. Three-Dimensional Cultures of Human Subcutaneous Adipose Tissue-Derived Progenitor Cells Based on RAD16-I Self-Assembling Peptide. Tissue Eng. Part C Methods 2016, 22, 113–124. [Google Scholar] [CrossRef]
- Semino, C.E.; Merok, J.R.; Crane, G.G.; Panagiotakos, G.; Zhang, S. Functional differentiation of hepatocyte-like spheroid structures from putative liver progenitor cells in three-dimensional peptide scaffolds. Differentiation 2003, 71, 262–270. [Google Scholar] [CrossRef] [Green Version]
- Genové, E.; Shen, C.; Zhang, S.; Semino, C.E. The effect of functionalized self-assembling peptide scaffolds on human aortic endothelial cell function. Biomaterials 2005, 26, 3341–3351. [Google Scholar] [CrossRef]
- Aloy-Reverté, C.; Moreno-Amador, J.L.; Nacher, M.; Montanya, E.; Semino, C.E. Use of RGD-Functionalized Sandwich Cultures to Promote Redifferentiation of Human Pancreatic Beta Cells After In Vitro Expansion. Tissue Eng. Part A. 2018, 24, 394–406. [Google Scholar] [CrossRef] [PubMed]
- Sankar, S.; O’Neill, K.; D’Arc, M.B.; Rebeca, F.; Buffier, M.; Aleksi, E.; Fan, M.; Matsuda, N.; Gil, E.S.; Spirio, L. Clinical Use of the Self-Assembling Peptide RADA16: A Review of Current and Future Trends in Biomedicine. Front. Bioeng. Biotechnol. 2021, 9, 465. [Google Scholar] [CrossRef] [PubMed]
- Olsen, J.V.; Ong, S.-E.; Mann, M. Trypsin Cleaves Exclusively C-terminal to Arginine and Lysine Residues. Mol Cell Proteom. 2004, 3, 608–614. [Google Scholar] [CrossRef] [Green Version]
- Lutolf, M.P.; Hubbell, J.A. Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nat. Biotechnol. 2005, 23, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Daura, X.; Bakowies, D.; Seebach, D.; Fleischhauer, J.; van Gunsteren, W.F.; Krüger, P. Circular dichroism spectra of β-peptides: Sensitivity to molecular structure and effects of motional averaging. Eur. Biophys. J. 2003, 32, 661–670. [Google Scholar] [CrossRef] [PubMed]
- Greenfield, N.J. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 2006, 1, 2876–2890. [Google Scholar] [CrossRef] [PubMed]
- Miroshnikova, Y.A.; Jorgens, D.M.; Spirio, L.; Auer, M.; Sarang-Sieminski, A.L.; Weaver, V.M. Engineering strategies to recapitulate epithelial morphogenesis within synthetic three-dimensional extracellular matrix with tunable mechanical properties. Phys. Biol. 2011, 8, 026013. [Google Scholar] [CrossRef] [Green Version]
- Song, H.; Cai, G.H.; Liang, J.; Ao, D.S.; Wang, H.; Yang, Z.H. Three-dimensional culture and clinical drug responses of a highly metastatic human ovarian cancer HO-8910PM cells in nanofibrous microenvironments of three hydrogel biomaterials. J. Nanobiotechnol. 2020, 18, 90. [Google Scholar] [CrossRef]
- Marí-Buyé, N.; Luque, T.; Navajas, D.; Semino, C.E. Development of a Three-Dimensional Bone-Like Construct in a Soft Self-Assembling Peptide Matrix. Tissue Eng. Part A 2013, 19, 870–881. [Google Scholar] [CrossRef] [Green Version]
- Recha-Sancho, L.; Semino, C.E. Heparin-based self-assembling peptide scaffold reestablish chondrogenic phenotype of expanded de-differentiated human chondrocytes. J. Biomed. Mater. Res. Part A 2016, 104, 1694–1706. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Zhou, T.; Tian, L.; Xia, Z.; Xu, F. Self-Assembling RADA16-I Peptide Hydrogel Scaffold Loaded with Tamoxifen for Breast Reconstruction. Biomed. Res. Int. 2017, 2017, 3656193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, C.; Wei, W.; Wang, Y.; Zhang, T.; Tang, Y.; Tang, F. Study of the interaction between self-assembling peptide and mangiferin and in vitro release of mangiferin from in situ hydrogel. Int. J. Nanomed. 2019, 14, 7447–7460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Briuglia, M.; Urquhart, A.J.; Lamprou, D.A. Sustained and controlled release of lipophilic drugs from a self-assembling amphiphilic peptide hydrogel. Int. J. Pharm. 2014, 474, 103–111. [Google Scholar] [CrossRef] [Green Version]
- Taka, E.; Karavasili, C.; Bouropoulos, N.; Moschakis, T.; Andreadis, D.D.D.; Zacharis, C.K.K.; Fatouros, D.G.G. Ocular Co-Delivery of Timolol and Brimonidine from a Self-Assembling Peptide Hydrogel for the Treatment of Glaucoma: In Vitro and Ex Vivo Evaluation. Pharmaceuticals 2020, 13, 126. [Google Scholar] [CrossRef]
- Mahmoudinobar, F.; Urban, J.M.; Su, Z.; Nilsson, B.L.; Dias, C.L. Thermodynamic Stability of Polar and Nonpolar Amyloid Fibrils. J. Chem. Theory Comput. 2019, 15, 3868–3874. [Google Scholar] [CrossRef]
- Cao, M.; Shen, Y.; Wang, Y.; Wang, X.; Li, D. Self-Assembly of Short Elastin-like Amphiphilic Peptides: Effects of temperature, molecular hydrophobicity and charge distribution. Molecules 2019, 24, 202. [Google Scholar] [CrossRef] [Green Version]
- Jalali, S.; Yang, Y.; Mahmoudinobar, F.; Singh, S.M.; Nilsson, B.L.; Dias, C. Using all-atom simulations in explicit solvent to study aggregation of amphipathic peptides into amyloid-like fibrils. J. Mol. Liq. 2022, 347, 118283. [Google Scholar] [CrossRef]
- Kisiday, J.; Jin, M.; Kurz, B.; Hung, H.; Semino, C.; Zhang, S.; Grodzinsky, A.J. Self-assembling peptide hydrogel fosters chondrocyte extracellular matrix production and cell division: Implications for cartilage tissue repair. Proc. Natl. Acad. Sci. USA 2002, 99, 9996–10001. [Google Scholar] [CrossRef] [Green Version]
- Urbic, T.; Najem, S.; Dias, C.L. Thermodynamic properties of amyloid fibrils in equilibrium. Biophys. Chem. 2017, 231, 155–160. [Google Scholar] [CrossRef]
- Wetzel, R. Kinetics and thermodynamics of amyloid fibril assembly. Acc. Chem. Res. 2006, 39, 671–679. [Google Scholar] [CrossRef] [PubMed]
- Gelain, F.; Luo, Z.; Zhang, S. Self-Assembling Peptide EAK16 and RADA16 Nanofiber Scaffold Hydrogel. Chem. Rev. 2020, 120, 13434–13460. [Google Scholar] [CrossRef] [PubMed]






| Peptide Fragment | Molecular Weight | Peak Presence in MALDI Spectra |
|---|---|---|
| AcN-RADARADARADARADA-CONH2 | 1712 | + |
| AcN-R | 216 | − |
| AcN-RADAR | 629 | + |
| AcN-RADARADAR | 1043 | + |
| AcN-RADARADARADAR | 1456 | + |
| ADA-CONH2 | 274 | − |
| ADAR | 431.4 | + |
| ADARADARADARADA-CONH2 | 1514 | − |
| ADARADARADA-CONH2 | 1102 | + |
| ADARADA-CONH2 | 688 | + |
| ADARADARADAR | 1258 | + |
| ADARADAR | 844 | + |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Genové, E.; Betriu, N.; Semino, C.E. β-Sheet to Random Coil Transition in Self-Assembling Peptide Scaffolds Promotes Proteolytic Degradation. Biomolecules 2022, 12, 411. https://0-doi-org.brum.beds.ac.uk/10.3390/biom12030411
Genové E, Betriu N, Semino CE. β-Sheet to Random Coil Transition in Self-Assembling Peptide Scaffolds Promotes Proteolytic Degradation. Biomolecules. 2022; 12(3):411. https://0-doi-org.brum.beds.ac.uk/10.3390/biom12030411
Chicago/Turabian StyleGenové, Elsa, Nausika Betriu, and Carlos E. Semino. 2022. "β-Sheet to Random Coil Transition in Self-Assembling Peptide Scaffolds Promotes Proteolytic Degradation" Biomolecules 12, no. 3: 411. https://0-doi-org.brum.beds.ac.uk/10.3390/biom12030411