Meroterpenoids Possibly Produced by a Bacterial Endosymbiont of the Tropical Basidiomycete Echinochaete brachypora
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fungal Materials
2.2. Cultivation of the Fungal Strain
2.3. Preparation of the Extracts
2.4. General Experimental Procedures
2.5. Isolation of Compounds from Strain MUCL 56080
2.6. DNA Extraction and PCR
2.7. Isolation Attempts of Fungal Endobacteria
2.8. Antimicrobial Assay
3. Results
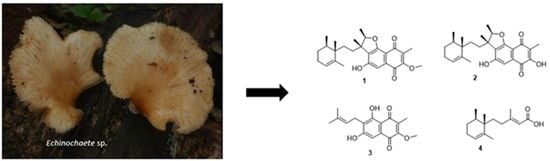
3.1. Structure Elucidation
3.2. Physicochemical Data for Compounds 1–4
3.3. Molecular Identification of MUCL 56080
3.4. Identification and Isolation of Endofungal Bacteria
3.5. Biological Activity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sandargo, B.; Chepkirui, C.; Cheng, T.; Chaverra-Muñoz, L.; Thongbai, B.; Stadler, M.; Hüttel, S. Biological and chemical diversity go hand in hand: Basidiomycota as source of new pharmaceuticals and agrochemicals. Biotechnol. Adv. 2019, 37, 107344. [Google Scholar] [CrossRef] [PubMed]
- Cheng, T.; Chepkirui, C.; Decock, C.; Matasyoh, J.C.; Stadler, M. Sesquiterpenes from an Eastern African medicinal mushroom belonging to the genus Sanghuangporus. J. Nat. Prod. 2019, 82, 1283–1291. [Google Scholar] [CrossRef] [PubMed]
- Chepkirui, C.; Cheng, T.; Sum, W.C.; Matasyoh, J.C.; Decock, C.; Praditya, D.F.; Wittstein, K.; Steinmann, E.; Stadler, M. Skeletocutins A-L: Antibacterial agents from the Kenyan wood-inhabiting basidiomycete, Skeletocutis sp. J. Agric. Food Chem. 2019, 67, 8468–8475. [Google Scholar] [CrossRef] [PubMed]
- Cheng, T.; Chepkirui, C.; Decock, C.; Matasyoh, J.C.; Stadler, M. Skeletocutins M–Q: Biologically active compounds from the fruiting bodies of the basidiomycete Skeletocutis sp. collected in Africa. Beilstein J. Org. Chem. 2019, 15, 2782–2789. [Google Scholar] [CrossRef] [Green Version]
- Chepkirui, C.; Matasyoh, J.C.; Decock, C.; Stadler, M. Two cytotoxic triterpenes from cultures of a Kenyan Laetiporus sp. (Basidiomycota). Phytochem. Lett. 2017, 20, 106–110. [Google Scholar] [CrossRef]
- Reid, D.A. New or interesting records of Australasian Basidiomycetes: V. Kew Bull. 1963, 17, 267–308. [Google Scholar] [CrossRef]
- Sotome, K.; Hattori, T.; Ota, Y.; Lee, S.S.; Vikineswary, S.; Abdullah, N.; Kakishima, M. Taxonomic study of Asian species of Echinochaete (Polyporaceae, Basidiomycota) and description of E. maximipora sp. nov. Mycol. Prog. 2009, 8, 123–132. [Google Scholar] [CrossRef]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar] [CrossRef]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef] [Green Version]
- Eden, P.A.; Schmidt, T.M.; Blakemore, R.P.; Pace, N.R. Phylogenetic analysis of Aquaspirillum magnetotacticum using polymerase chain reaction-amplified 16s rRNA-specific DNA. Int. Union Microbiol. Soc. 1991, 41, 324–325. [Google Scholar] [CrossRef]
- Muyzer, G.; De Waal, E.C.; Uitterlinden, A.G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 1993, 59, 695–700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Khosravi Babadi, Z.; Ebrahimipour, G.; Wink, J.; Narmani, A.; Risdian, C. Isolation and identification of Streptomyces sp. Act4Zk, a good producer of staurosporine and some derivatives. Lett. Appl. Microbiol. 2021, 72, 206–218. [Google Scholar] [CrossRef] [PubMed]
- Becker, K.; Wessel, A.C.; Luangsa-Ard, J.J.; Stadler, M. Viridistratins A-C, antimicrobial and cytotoxic benzo fluoranthenes from stromata of Annulohypoxylon viridistratum (Hypoxylaceae, Ascomycota). Biomolecules 2020, 10, 805. [Google Scholar] [CrossRef]
- Kemkuignou, B.M.; Treiber, L.; Zeng, H.; Schrey, H.; Schobert, R.; Stadler, M. Macrooxazoles a–d, new 2,5-disubstituted oxazole-4-carboxylic acid derivatives from the plant pathogenic fungus Phoma macrostoma. Molecules 2020, 25, 5497. [Google Scholar] [CrossRef]
- Hassan, K.; Kemkuignou, B.M.; Stadler, M. Two new triterpenes from basidiomata of the medicinal and edible mushroom, Laetiporus sulphureus. Molecules 2021, 26, 7090. [Google Scholar] [CrossRef]
- Hardt, I.H.; Jensen, P.R.; Fenical, W. Neomarinone, and new cytotoxic marinone derivatives, produced by a marine filamentous bacterium (Actinomycetales). Tetrahedron Lett. 2000, 41, 2073–2076. [Google Scholar] [CrossRef]
- Kalaitzis, J.A.; Hamano, Y.; Nilsen, G.; Moore, B.S. Biosynthesis and structural revision of neomarinone. Org. Lett. 2003, 5, 4449–4452. [Google Scholar] [CrossRef]
- Charan, R.D.; Schlingmann, G.; Bernan, V.S.; Feng, X.; Carter, G.T. Fumaquinone, a new prenylated naphthoquinone from Streptomyces fumanus. J. Antibiot. 2005, 58, 271–274. [Google Scholar] [CrossRef]
- Masuka, A.; Decock, C.; Mossebo, D.; Ryvarden, L. Poroid Fungi of Africa; Fungiflora: Oslo, Norway, 2022; ISBN 9788290724639. [Google Scholar]
- Yarza, P.; Yilmaz, P.; Pruesse, E.; Glöckner, F.O.; Ludwig, W.; Schleifer, K.-H.; Whitman, W.B.; Euzéby, J.; Amann, R.; Rosselló-Móra, R. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Rev. Microbiol. 2014, 12, 635–645. [Google Scholar] [CrossRef]
- Song, Y.; Huang, H.; Chen, Y.; Ding, J.; Zhang, Y.; Sun, A.; Zhang, W.; Ju, J. Cytotoxic and antibacterial marfuraquinocins from the deep south china sea-derived Streptomyces niveus scsio 3406. J. Nat. Prod. 2013, 76, 2263–2268. [Google Scholar] [CrossRef]
- Jenner, M.; Jian, X.; Dashti, Y.; Masschelein, J.; Hobson, C.; Roberts, D.M.; Jones, C.; Harris, S.; Parkhill, J.; Raja, H.A.; et al. An unusual: Burkholderia gladioli double chain-initiating nonribosomal peptide synthetase assembles ″fungal″ icosalide antibiotics. Chem. Sci. 2019, 10, 5489–5494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujishima, M.; Kodama, Y. Endosymbionts in Paramecium. Eur. J. Protistol. 2012, 48, 124–137. [Google Scholar] [CrossRef] [PubMed]
- Schüβler, A.; Schwarzott, D.; Walker, C. A new fungal phylum, the Glomeromycota: Phylogeny and evolution. Mycol. Res. 2001, 105, 1413–1421. [Google Scholar] [CrossRef] [Green Version]
- Cruz, A.F.; Horii, S.; Ochiai, S.; Yasuda, A.; Ishii, T. Isolation and analysis of bacteria associated with spores of Gigaspora margarita. J. Appl. Microbiol. 2008, 104, 1711–1717. [Google Scholar] [CrossRef] [PubMed]
- Bianciotto, V.; Lumini, E.; Bonfante, P.; Vandamme, P. “Candidatus Glomeribacter gigasporarum” gen. nov., sp. nov., an endosymbiont of arbuscular mycorrhizal fungi. Int. J. Syst. Evol. Microbiol. 2003, 53, 121–124. [Google Scholar] [CrossRef]
- Ryan, M.P.; Adley, C.C. Ralstonia spp.: Emerging global opportunistic pathogens. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 291–304. [Google Scholar] [CrossRef] [Green Version]
- Guo, Y.; Narisawa, K. Fungus-bacterium symbionts promote plant health and performance. Microbes Environ. 2018, 33, 239–241. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Qiu, S. Phylogenomic analysis of the genus Ralstonia based on 686 single-copy genes. Antonie Leeuwenhoek 2016, 109, 71–82. [Google Scholar] [CrossRef]
- Ryan, M.P.; Pembroke, J.T.; Adley, C.C. Ralstonia pickettii in environmental biotechnology: Potential and applications. J. Appl. Microbiol. 2007, 103, 754–764. [Google Scholar] [CrossRef] [Green Version]
- Ferro, P.; Vaz-Moreira, I.; Manaia, C.M. Association between gentamicin resistance and stress tolerance in water isolates of Ralstonia pickettii and R. mannitolilytica. Folia Microbiol. 2019, 64, 63–72. [Google Scholar] [CrossRef] [Green Version]
- Hassan, E.A.; Balabel, N.M.; Ahmed, A.E.; Eid, N.A.; Ramadan, E.M. Relationship between Ralstonia solanacearum and bioagents recovered from different habitats. Int. J. Sci. Eng. Res. 2017, 8, 91–104. [Google Scholar]
- Bonfante, P. Plants, mycorrhizal fungi and endobacteria: A dialog among cells and genomes. Biol. Bull. 2003, 204, 215–220. [Google Scholar] [CrossRef]
- Bertaux, J.; Schmid, M.; Prevost-Boure, N.C.; Churin, J.L.; Hartmann, A.; Garbaye, J.; Frey-Klett, P. In situ identification of intracellular bacteria related to Paenibacillus spp. in the mycelium of the ectomycorrhizal fungus Laccaria bicolor S238N. Appl. Environ. Microbiol. 2003, 69, 4243–4248. [Google Scholar] [CrossRef] [Green Version]
- Aslani, M.A.; Harighi, B.; Abdollahzadeh, J. Screening of endofungal bacteria isolated from wild growing mushrooms as potential biological control agents against brown blotch and internal stipe necrosis diseases of Agaricus bisporus. Biol. Control 2018, 119, 20–26. [Google Scholar] [CrossRef]
- Sharma, M.; Schmid, M.; Rothballer, M.; Hause, G.; Zuccaro, A.; Imani, J.; Kämpfer, P.; Domann, E.; Schäfer, P.; Hartmann, A.; et al. Detection and identification of bacteria intimately associated with fungi of the order Sebacinales. Cell. Microbiol. 2008, 10, 2235–2246. [Google Scholar] [CrossRef]
- Partida-Martinez, L.P.; Hertweck, C. Pathogenic fungus harbours endosymbiotic bacteria for toxin production. Nature 2005, 437, 884–888. [Google Scholar] [CrossRef]
- Partida-Martinez, L.P.; De Looß, C.F.; Ishida, K.; Ishida, M.; Roth, M.; Buder, K.; Hertweck, C. Rhizonin, the first mycotoxin isolated from the Zygomycota, is not a fungal metabolite but is produced by bacterial endosymbionts. Appl. Environ. Microbiol. 2007, 73, 793–797. [Google Scholar] [CrossRef] [Green Version]
- Mosse, B. Honey-coloured, sessile endogone spores: II. Changes in fine structure during spore development. Arch. Mikrobiol. 1970, 74, 129–145. [Google Scholar] [CrossRef]
- Guo, H.; Glaeser, S.P.; Alabid, I.; Imani, J.; Haghighi, H.; Kämpfer, P.; Kogel, K.-H. The abundance of endofungal bacterium Rhizobium radiobacter (syn. Agrobacterium tumefaciens) increases in its fungal host Piriformospora indica during the tripartite sebacinalean symbiosis with higher plants. Front. Microbiol. 2017, 8, 629. [Google Scholar] [CrossRef]
- Kobayashi, D.Y.; Crouch, J.A. Bacterial/fungal interactions: From pathogens to mutualistic endosymbionts. Annu. Rev. Phytopathol. 2009, 47, 63–82. [Google Scholar] [CrossRef] [Green Version]
- Alabid, I.; Glaeser, S.P.; Kogel, K.-H. Endofungal bacteria increase fitness of their host fungi and impact their association with crop plants. Curr. Issues Mol. Biol. 2019, 30, 59–74. [Google Scholar] [CrossRef]
- Pent, M.; Bahram, M.; Põldmaa, K. Fruitbody chemistry underlies the structure of endofungal bacterial communities across fungal guilds and phylogenetic groups. ISME J. 2020, 14, 2131–2141. [Google Scholar] [CrossRef]
- Lackner, G.; Hertweck, C. Impact of endofungal bacteria on infection biology, food safety, and drug development. PLoS Pathog. 2011, 7, e1002096. [Google Scholar] [CrossRef] [Green Version]
- Lackner, G.; Partida-Martinez, L.P.; Hertweck, C. Endofungal bacteria as producers of mycotoxins. Trends Microbiol. 2009, 17, 570–576. [Google Scholar] [CrossRef]
- Spraker, J.E.; Sanchez, L.M.; Lowe, T.M.; Dorrestein, P.C.; Keller, N.P. Ralstonia solanacearum lipopeptide induces chlamydospore development in fungi and facilitates bacterial entry into fungal tissues. ISME J. 2016, 10, 2317–2330. [Google Scholar] [CrossRef] [Green Version]



| 1 | 2 | 4 | ||||
|---|---|---|---|---|---|---|
| No. | 13C/HSQC | 1H | 13C/HSQC | 1H | 13C/HSQC | 1H |
| 1. | 157.6, C | 153.7, C | 167.8, C | |||
| 2. | 181.4, C | 181.5, C | 115.9, CH | 5.69 (q), J = 1.1Hz | ||
| 3. | 134.7, C | 132.7, C | 161.7, C | |||
| 4. | 108.9, CH | 7.14 (s) | 108.6, CH | 1.17 (s) | 36.4, CH2 | 1.81 (m), 2.13 (m) |
| 5. | 158.7, C | 158.2, C | 35.4, CH2 | 1.55 (m), 1.61 (m) | ||
| 6. | 128.1, C | 129.1, C | 41.3, C | |||
| 7. | 161.7, C | 161.8, C | 34.2, CH | 1.75 (m) | ||
| 8. | 109.8, C | 109.9, C | 27.8, CH2 | 1.47 (m) | ||
| 9. | 183.5, C | 183.5, C | 26.2, CH2 | 1.92 (m), 1.98 (m) | ||
| 10. | 133.3, C | 121.3, C | 125.4, CH | 5.45 (m) | ||
| 11. | 16.2, CH3 | 1.38 (d) J = 6.7 Hz | 16.2, CH3 | 1.37 (d) J = 6.5 Hz | 139.9, C | |
| 12. | 88.4, CH | 4.76 (q) J = 6.6 Hz | 88.2, CH | 4.76 (q) J = 6.6 Hz | 19.4, CH3 | 1.63 (s) |
| 13. | 47.4, C | 47.5, C | 16.2, CH3 | 0.89, (m) | ||
| 14. | 33.2, CH2 | 1.66 (m) | 33.1, CH2 | 1.67 (m) | 21.3, CH3 | 0.90, (s) |
| 15. | 32.2, CH2 | 1.29 (m), 1.46 (m) | 32.1, CH2 | 1.30 (m), 1.46 (m) | 18.9, CH3 | 2.16 (d), J = 1.2 Hz |
| 16. | 40.9, C | 40.9, C | ||||
| 17. | 34.1, CH | 1.79 (m) | 34.1, CH | 1.78 (m) | ||
| 18. | 27.8, CH2 | 1.39 (m), 1.46 (m) | 27.8, CH2 | 1.43 (m) | ||
| 19. | 26.0, CH2 | 1.88 (m), 1.93 (m) | 26.0, CH2 | 1.9 0 (m) | ||
| 20. | 124.4, CH | 5.36 (m) | 124.6, CH | 5.36 (m) | ||
| 21. | 140.4, C | 140.4, C | ||||
| 22. | 19.4, CH3 | 1.59 (s) | 19.4, CH3 | 1.58 (s) | ||
| 23. | 16.2, CH3 | 0.86 (d) J = 7.1 Hz | 16.2, CH3 | 0.86 (d) J = 6.9 Hz | ||
| 24. | 21.7, CH3 | 0.82 (s) | 21.7, CH3 | 0.82 (s) | ||
| 25. | 20.2, CH3 | 1.31 (s) | 20.1, CH3 | 1.31 (s) | ||
| 26. | 9.4, CH3 | 1.96 (s) | 8.7, CH3 | 1.95 (s) | ||
| OCH3 | 60.8, CH3 | 3.98 (s) |
| MIC (μg/mL) | |||||
|---|---|---|---|---|---|
| Test Organism | 1 | 2 | 3 | 4 | Positive Control |
| Bacillus subtilis DSM10 | 4.6 | 4.6 | 4.6 | - | 2.3 a |
| Escherichia coli DSM 498 | - | - | - | - | 2.3 a |
| Mucor plumbeus MUCL 49355 | - | - | - | - | 9.4 b |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hassan, K.; Chepkirui, C.; Llanos-López, N.A.; Matasyoh, J.C.; Decock, C.; Marin-Felix, Y.; Stadler, M. Meroterpenoids Possibly Produced by a Bacterial Endosymbiont of the Tropical Basidiomycete Echinochaete brachypora. Biomolecules 2022, 12, 755. https://0-doi-org.brum.beds.ac.uk/10.3390/biom12060755
Hassan K, Chepkirui C, Llanos-López NA, Matasyoh JC, Decock C, Marin-Felix Y, Stadler M. Meroterpenoids Possibly Produced by a Bacterial Endosymbiont of the Tropical Basidiomycete Echinochaete brachypora. Biomolecules. 2022; 12(6):755. https://0-doi-org.brum.beds.ac.uk/10.3390/biom12060755
Chicago/Turabian StyleHassan, Khadija, Clara Chepkirui, Natalia Andrea Llanos-López, Josphat C. Matasyoh, Cony Decock, Yasmina Marin-Felix, and Marc Stadler. 2022. "Meroterpenoids Possibly Produced by a Bacterial Endosymbiont of the Tropical Basidiomycete Echinochaete brachypora" Biomolecules 12, no. 6: 755. https://0-doi-org.brum.beds.ac.uk/10.3390/biom12060755