Neuroproteomic Analysis after SARS-CoV-2 Infection Reveals Overrepresented Neurodegeneration Pathways and Disrupted Metabolic Pathways
Abstract
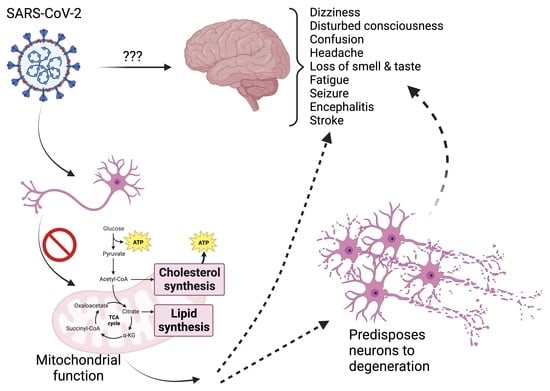
:1. Introduction
2. Materials and Methods
2.1. Generation of Induced Pluripotent Stem Cell-Derived Human Neurons
2.2. Generation of Induced Pluripotent Stem Cell-Derived Human Astrocytes
2.3. SARS-CoV-2 Production and Infection of i3Ns and iAs
2.4. Assessment of SARS-CoV-2 Infection and Replication
2.5. Neuroproteomic Analysis of SARS-CoV-2 Infected Human Neurons
2.6. Label-Free Proteomic Analysis of SARS-CoV-2 Infected Neurons
2.7. Statistical Analysis
3. Results
3.1. SARS-CoV-2 Infects iPSC-Derived Human Neurons and Astrocytes
3.2. SARS-CoV-2 Does Not Replicate within Human Neurons
3.3. SARS-CoV-2 Infection in Neurons Shows Distinct Changes in Neuronal Proteome
3.4. SARS-CoV-2 Infection Affects Apoptotic and Metabolic Pathways in Neurons
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef]
- Mao, L.; Jin, H.; Wang, M.; Hu, Y.; Chen, S.; He, Q.; Chang, J.; Hong, C.; Zhou, Y.; Wang, D.; et al. Neurologic Manifestations of Hospitalized Patients With Coronavirus Disease 2019 in Wuhan, China. JAMA Neurol. 2020, 77, 683–690. [Google Scholar] [CrossRef] [PubMed]
- Helms, J.; Kremer, S.; Merdji, H.; Clere-Jehl, R.; Schenck, M.; Kummerlen, C.; Collange, O.; Boulay, C.; Fafi-Kremer, S.; Ohana, M.; et al. Neurologic Features in Severe SARS-CoV-2 Infection. N. Engl. J. Med. 2020, 382, 2268–2270. [Google Scholar] [CrossRef] [PubMed]
- Xydakis, M.S.; Dehgani-Mobaraki, P.; Holbrook, E.H.; Geisthoff, U.W.; Bauer, C.; Hautefort, C.; Herman, P.; Manley, G.T.; Lyon, D.M.; Hopkins, C. Smell and taste dysfunction in patients with COVID-19. Lancet Infect. Dis. 2020, 20, 1015–1016. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Pan, M.; Xiao, Z.; Xu, X. Neurological manifestations of the coronavirus (SARS-CoV-2) pandemic 2019-2020. J. Neurol. Neurosurg. Psychiatry 2020, 91, 669–670. [Google Scholar] [CrossRef]
- Ye, M.; Ren, Y.; Lv, T. Encephalitis as a clinical manifestation of COVID-19. Brain Behav. Immun. 2020, 88, 945–946. [Google Scholar] [CrossRef] [PubMed]
- Oxley, T.J.; Mocco, J.; Majidi, S.; Kellner, C.P.; Shoirah, H.; Singh, I.P.; De Leacy, R.A.; Shigematsu, T.; Ladner, T.R.; Yaeger, K.A.; et al. Large-Vessel Stroke as a Presenting Feature of COVID-19 in the Young. N. Engl. J. Med. 2020, 382, e60. [Google Scholar] [CrossRef]
- Tunc, A.; Unlubas, Y.; Alemdar, M.; Akyuz, E. Coexistence of COVID-19 and acute ischemic stroke report of four cases. J. Clin. Neurosci. 2020, 77, 227–229. [Google Scholar] [CrossRef]
- Planchuelo-Gomez, A.; Garcia-Azorin, D.; Guerrero, A.L.; Rodriguez, M.; Aja-Fernandez, S.; de Luis-Garcia, R. Structural brain changes in patients with persistent headache after COVID-19 resolution. J. Neurol. 2023, 270, 13–31. [Google Scholar] [CrossRef]
- Ding, Y.; Wang, H.; Shen, H.; Li, Z.; Geng, J.; Han, H.; Cai, J.; Li, X.; Kang, W.; Weng, D.; et al. The clinical pathology of severe acute respiratory syndrome (SARS): A report from China. J. Pathol. 2003, 200, 282–289. [Google Scholar] [CrossRef]
- Paniz-Mondolfi, A.; Bryce, C.; Grimes, Z.; Gordon, R.E.; Reidy, J.; Lednicky, J.; Sordillo, E.M.; Fowkes, M. Central Nervous System Involvement by Severe Acute Respiratory Syndrome Coronavirus -2 (SARS-CoV-2). J. Med. Virol. 2020, 92, 699–702. [Google Scholar] [CrossRef]
- Lee, M.H.; Perl, D.P.; Nair, G.; Li, W.; Maric, D.; Murray, H.; Dodd, S.J.; Koretsky, A.P.; Watts, J.A.; Cheung, V.; et al. Microvascular Injury in the Brains of Patients with COVID-19. N. Engl. J. Med. 2021, 384, 481–483. [Google Scholar] [CrossRef]
- Harapan, B.N.; Yoo, H.J. Neurological symptoms, manifestations, and complications associated with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease 19 (COVID-19). J. Neurol. 2021, 268, 3059–3071. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.M.; White, N.; Premraj, L.; Battaglini, D.; Fanning, J.; Suen, J.; Bassi, G.L.; Fraser, J.; Robba, C.; Griffee, M.; et al. Neurological manifestations of COVID-19 in adults and children. Brain 2023, 146, 1648–1661. [Google Scholar] [CrossRef] [PubMed]
- Massimo, M.; Barelli, C.; Moreno, C.; Collesi, C.; Holloway, R.K.; Crespo, B.; Zentilin, L.; Williams, A.; Miron, V.E.; Giacca, M.; et al. Haemorrhage of human foetal cortex associated with SARS-CoV-2 infection. Brain 2023, 146, 1175–1185. [Google Scholar] [CrossRef] [PubMed]
- Taquet, M.; Geddes, J.R.; Husain, M.; Luciano, S.; Harrison, P.J. 6-month neurological and psychiatric outcomes in 236 379 survivors of COVID-19: A retrospective cohort study using electronic health records. Lancet Psychiatry 2021, 8, 416–427. [Google Scholar] [CrossRef]
- Douaud, G.; Lee, S.; Alfaro-Almagro, F.; Arthofer, C.; Wang, C.; McCarthy, P.; Lange, F.; Andersson, J.L.R.; Griffanti, L.; Duff, E.; et al. SARS-CoV-2 is associated with changes in brain structure in UK Biobank. Nature 2022, 604, 697–707. [Google Scholar] [CrossRef]
- Du, Y.; Zhao, W.; Huang, S.; Huang, Y.; Chen, Y.; Zhang, H.; Guo, H.; Liu, J. Two-year follow-up of brain structural changes in patients who recovered from COVID-19: A prospective study. Psychiatry Res. 2023, 319, 114969. [Google Scholar] [CrossRef]
- Zhou, S.; Wei, T.; Liu, X.; Liu, Y.; Song, W.; Que, X.; Xing, Y.; Wang, Z.; Tang, Y. Causal effects of COVID-19 on structural changes in specific brain regions: A Mendelian randomization study. BMC Med. 2023, 21, 261. [Google Scholar] [CrossRef]
- Díez-Cirarda, M.; Yus, M.; Gómez-Ruiz, N.; Polidura, C.; Gil-Martínez, L.; Delgado-Alonso, C.; Jorquera, M.; Gómez-Pinedo, U.; Matias-Guiu, J.; Arrazola, J.; et al. Multimodal neuroimaging in post-COVID syndrome and correlation with cognition. Brain 2023, 146, 2142–2152. [Google Scholar] [CrossRef]
- Yang, A.C.; Kern, F.; Losada, P.M.; Agam, M.R.; Maat, C.A.; Schmartz, G.P.; Fehlmann, T.; Stein, J.A.; Schaum, N.; Lee, D.P.; et al. Dysregulation of brain and choroid plexus cell types in severe COVID-19. Nature 2021, 595, 565–571. [Google Scholar] [CrossRef] [PubMed]
- Xu, E.; Xie, Y.; Al-Aly, Z. Long-term neurologic outcomes of COVID-19. Nat. Med. 2022, 28, 2406–2415. [Google Scholar] [CrossRef] [PubMed]
- Taquet, M.; Sillett, R.; Zhu, L.; Mendel, J.; Camplisson, I.; Dercon, Q.; Harrison, P.J. Neurological and psychiatric risk trajectories after SARS-CoV-2 infection: An analysis of 2-year retrospective cohort studies including 1,284,437 patients. Lancet Psychiatry 2022, 9, 815–827. [Google Scholar] [CrossRef] [PubMed]
- Rass, V.; Beer, R.; Schiefecker, A.J.; Lindner, A.; Kofler, M.; Ianosi, B.A.; Mahlknecht, P.; Heim, B.; Peball, M.; Carbone, F.; et al. Neurological outcomes 1 year after COVID-19 diagnosis: A prospective longitudinal cohort study. Eur. J. Neurol. 2022, 29, 1685–1696. [Google Scholar] [CrossRef]
- Khullar, S.; Wang, D. Predicting brain-regional gene regulatory networks from multi-omics for Alzheimer’s disease phenotypes and COVID-19 severity. Hum. Mol. Genet. 2023, 32, 1797–1813. [Google Scholar] [CrossRef]
- Baranova, A.; Cao, H.; Teng, S.; Su, K.P.; Zhang, F. Shared genetics and causal associations between COVID-19 and multiple sclerosis. J. Med. Virol. 2023, 95, e28431. [Google Scholar] [CrossRef]
- Mavrikaki, M.; Lee, J.D.; Solomon, I.H.; Slack, F.J. Severe COVID-19 is associated with molecular signatures of aging in the human brain. Nat. Aging 2022, 2, 1130–1137. [Google Scholar] [CrossRef] [PubMed]
- Strong, M.J. SARS-CoV-2, aging, and Post-COVID-19 neurodegeneration. J. Neurochem. 2023, 165, 115–130. [Google Scholar] [CrossRef]
- Fu, Y.; Guo, Z.; Wang, Y.; Zhang, H.; Zhang, F.; Xu, Z.; Shen, X.; Roppongi, R.T.; Mo, S.; Gu, W.; et al. Single-nucleus RNA sequencing reveals the shared mechanisms inducing cognitive impairment between COVID-19 and Alzheimer’s disease. Front. Immunol. 2022, 13, 967356. [Google Scholar] [CrossRef]
- Chen, Y.; Yao, H.; Zhang, N.; Wu, J.; Gao, S.; Guo, J.; Lu, X.; Cheng, L.; Luo, R.; Liang, X.; et al. Proteomic Analysis Identifies Prolonged Disturbances in Pathways Related to Cholesterol Metabolism and Myocardium Function in the COVID-19 Recovery Stage. J. Proteome Res. 2021, 20, 3463–3474. [Google Scholar] [CrossRef]
- Crunfli, F.; Carregari, V.C.; Veras, F.P.; Silva, L.S.; Nogueira, M.H.; Antunes, A.; Vendramini, P.H.; Valenca, A.G.F.; Brandao-Teles, C.; Zuccoli, G.D.S.; et al. Morphological, cellular, and molecular basis of brain infection in COVID-19 patients. Proc. Natl. Acad. Sci. USA 2022, 119, e2200960119. [Google Scholar] [CrossRef]
- De Oliveira, L.G.; de Souza Angelo, Y.; Yamamoto, P.; Carregari, V.C.; Crunfli, F.; Reis-de-Oliveira, G.; Costa, L.; Vendramini, P.H.; Duque, E.A.; Dos Santos, N.B.; et al. SARS-CoV-2 infection impacts carbon metabolism and depends on glutamine for replication in Syrian hamster astrocytes. J. Neurochem. 2022, 163, 113–132. [Google Scholar] [CrossRef] [PubMed]
- Iosef, C.; Knauer, M.J.; Nicholson, M.; Van Nynatten, L.R.; Cepinskas, G.; Draghici, S.; Han, V.K.M.; Fraser, D.D. Plasma proteome of Long-COVID patients indicates HIF-mediated vasculo-proliferative disease with impact on brain and heart function. J. Transl. Med. 2023, 21, 377. [Google Scholar] [CrossRef] [PubMed]
- Kettunen, P.; Lesnikova, A.; Räsänen, N.; Ojha, R.; Palmunen, L.; Laakso, M.; Lehtonen, Š.; Kuusisto, J.; Pietiläinen, O.; Saber, S.H.; et al. SARS-CoV-2 Infection of Human Neurons Is TMPRSS2 Independent, Requires Endosomal Cell Entry, and Can Be Blocked by Inhibitors of Host Phosphoinositol-5 Kinase. J. Virol. 2023, 97, e0014423. [Google Scholar] [CrossRef]
- Maity, S.; Mayer, M.G.; Shu, Q.; Linh, H.; Bao, D.; Blair, R.V.; He, Y.; Lyon, C.J.; Hu, T.Y.; Fischer, T.; et al. Cerebrospinal Fluid Protein Markers Indicate Neuro-Damage in SARS-CoV-2-Infected Nonhuman Primates. Mol. Cell. Proteom. 2023, 22, 100523. [Google Scholar] [CrossRef]
- Reinhold, D.; Farztdinov, V.; Yan, Y.; Meisel, C.; Sadlowski, H.; Kuhn, J.; Perschel, F.H.; Endres, M.; Duzel, E.; Vielhaber, S.; et al. The brain reacting to COVID-19: Analysis of the cerebrospinal fluid proteome, RNA and inflammation. J. Neuroinflammation 2023, 20, 30. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Qin, W.; Guo, Z.; Li, X.; Li, F.; Wang, X. Application of weighted gene co-expression network and immune infiltration for explorations of key genes in the brain of elderly COVID-19 patients. Front. Immunol. 2023, 14, 1157179. [Google Scholar] [CrossRef]
- Finlay, J.B.; Brann, D.H.; Abi Hachem, R.; Jang, D.W.; Oliva, A.D.; Ko, T.; Gupta, R.; Wellford, S.A.; Moseman, E.A.; Jang, S.S.; et al. Persistent post-COVID-19 smell loss is associated with immune cell infiltration and altered gene expression in olfactory epithelium. Sci. Transl. Med. 2022, 14, eadd0484. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Kruger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef]
- Baggen, J.; Jacquemyn, M.; Persoons, L.; Vanstreels, E.; Pye, V.E.; Wrobel, A.G.; Calvaresi, V.; Martin, S.R.; Roustan, C.; Cronin, N.B.; et al. TMEM106B is a receptor mediating ACE2-independent SARS-CoV-2 cell entry. Cell 2023, 186, 3427–3442.e22. [Google Scholar] [CrossRef]
- Zhang, Q.; Xiang, R.; Huo, S.; Zhou, Y.; Jiang, S.; Wang, Q.; Yu, F. Molecular mechanism of interaction between SARS-CoV-2 and host cells and interventional therapy. Signal Transduct. Target. Ther. 2021, 6, 233. [Google Scholar] [CrossRef] [PubMed]
- Finkel, Y.; Gluck, A.; Nachshon, A.; Winkler, R.; Fisher, T.; Rozman, B.; Mizrahi, O.; Lubelsky, Y.; Zuckerman, B.; Slobodin, B.; et al. SARS-CoV-2 uses a multipronged strategy to impede host protein synthesis. Nature 2021, 594, 240–245. [Google Scholar] [CrossRef] [PubMed]
- Filbin, M.R.; Mehta, A.; Schneider, A.M.; Kays, K.R.; Guess, J.R.; Gentili, M.; Fenyves, B.G.; Charland, N.C.; Gonye, A.L.K.; Gushterova, I.; et al. Longitudinal proteomic analysis of severe COVID-19 reveals survival-associated signatures, tissue-specific cell death, and cell-cell interactions. Cell Rep. Med. 2021, 2, 100287. [Google Scholar] [CrossRef]
- Schweizer, L.; Schaller, T.; Zwiebel, M.; Karayel, O.; Muller-Reif, J.B.; Zeng, W.F.; Dintner, S.; Nordmann, T.M.; Hirschbuhl, K.; Markl, B.; et al. Quantitative multiorgan proteomics of fatal COVID-19 uncovers tissue-specific effects beyond inflammation. EMBO Mol. Med. 2023, 15, e17459. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Miao, F.; Lv, S.; Li, L.; Wei, F.; Hou, L.; Sun, R.; Li, W.; Zhang, J.; Zhang, C.; et al. Proteomic and Metabolomic Characterization of SARS-CoV-2-Infected Cynomolgus Macaque at Early Stage. Front. Immunol. 2022, 13, 954121. [Google Scholar] [CrossRef]
- Basak, I.; Hansen, R.A.; Ward, M.E.; Hughes, S.M. Deficiency of the Lysosomal Protein CLN5 Alters Lysosomal Function and Movement. Biomolecules 2021, 11, 1412. [Google Scholar] [CrossRef] [PubMed]
- Fernandopulle, M.S.; Prestil, R.; Grunseich, C.; Wang, C.; Gan, L.; Ward, M.E. Transcription Factor-Mediated Differentiation of Human iPSCs into Neurons. Curr. Protoc. Cell Biol. 2018, 79, e51. [Google Scholar]
- Tian, R.; Gachechiladze, M.A.; Ludwig, C.H.; Laurie, M.T.; Hong, J.Y.; Nathaniel, D.; Prabhu, A.V.; Fernandopulle, M.S.; Patel, R.; Abshari, M.; et al. CRISPR Interference-Based Platform for Multimodal Genetic Screens in Human iPSC-Derived Neurons. Neuron 2019, 104, 239–255.e12. [Google Scholar] [CrossRef]
- Peppercorn, K.; Kleffmann, T.; Jones, O.; Hughes, S.; Tate, W. Secreted Amyloid Precursor Protein Alpha, a Neuroprotective Protein in the Brain Has Widespread Effects on the Transcriptome and Proteome of Human Inducible Pluripotent Stem Cell-Derived Glutamatergic Neurons Related to Memory Mechanisms. Front. Neurosci. 2022, 16, 858524. [Google Scholar] [CrossRef]
- Canals, I.; Ginisty, A.; Quist, E.; Timmerman, R.; Fritze, J.; Miskinyte, G.; Monni, E.; Hansen, M.G.; Hidalgo, I.; Bryder, D.; et al. Rapid and efficient induction of functional astrocytes from human pluripotent stem cells. Nat. Methods 2018, 15, 693–696. [Google Scholar] [CrossRef]
- Brozzi, F.; Arcuri, C.; Giambanco, I.; Donato, R. S100B Protein Regulates Astrocyte Shape and Migration via Interaction with Src Kinase: Implications for Astrocyte Development, Activation, and Tumor Growth. J. Biol. Chem. 2009, 284, 8797–8811. [Google Scholar] [CrossRef]
- Harfoot, R.; Lawley, B.; Hernández, L.C.; Kuang, J.; Grant, J.; Treece, J.M.; LeQueux, S.; Day, R.; Jack, S.; Stanton, J.L.; et al. Characterization of the First SARS-CoV-2 Isolates from Aotearoa New Zealand as Part of a Rapid Response to the COVID-19 Pandemic. Viruses 2022, 14, 366. [Google Scholar] [CrossRef] [PubMed]
- Reed, L.J.; Muench, H. A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- Basak, I.; Bhatlekar, S.; Manne, B.K.; Stoller, M.; Hugo, S.; Kong, X.; Ma, L.; Rondina, M.T.; Weyrich, A.S.; Edelstein, L.C.; et al. miR-15a-5p regulates expression of multiple proteins in the megakaryocyte GPVI signaling pathway. J. Thromb. Haemost. 2019, 17, 511–524. [Google Scholar] [CrossRef] [PubMed]
- Best, H.L.; Neverman, N.J.; Wicky, H.E.; Mitchell, N.L.; Leitch, B.; Hughes, S.M. Characterisation of early changes in ovine CLN5 and CLN6 Batten disease neural cultures for the rapid screening of therapeutics. Neurobiol. Dis. 2017, 100, 62–74. [Google Scholar] [CrossRef]
- Lawley, B.; Grant, J.; Harfoot, R.; Treece, J.M.; Day, R.; Hernández, L.C.; Stanton, J.L.; Ussher, J.E.; Quiñones-Mateu, M.E. Rapid Response to SARS-CoV-2 in Aotearoa New Zealand: Implementation of a Diagnostic Test and Characterization of the First COVID-19 Cases in the South Island. Viruses 2021, 13, 2222. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Xie, Z.; Bailey, A.; Kuleshov, M.V.; Clarke, D.J.B.; Evangelista, J.E.; Jenkins, S.L.; Lachmann, A.; Wojciechowicz, M.L.; Kropiwnicki, E.; Jagodnik, K.M.; et al. Gene Set Knowledge Discovery with Enrichr. Curr. Protoc. 2021, 1, e90. [Google Scholar] [CrossRef] [PubMed]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Silva, J.C.; Gorenstein, M.V.; Li, G.Z.; Vissers, J.P.; Geromanos, S.J. Absolute quantification of proteins by LCMSE: A virtue of parallel MS acquisition. Mol. Cell. Proteom. 2006, 5, 144–156. [Google Scholar] [CrossRef]
- Li, N.; Hui, H.; Bray, B.; Gonzalez, G.M.; Zeller, M.; Anderson, K.G.; Knight, R.; Smith, D.; Wang, Y.; Carlin, A.F.; et al. METTL3 regulates viral m6A RNA modification and host cell innate immune responses during SARS-CoV-2 infection. Cell Rep. 2021, 35, 109091. [Google Scholar] [CrossRef]
- Ramani, A.; Muller, L.; Ostermann, P.N.; Gabriel, E.; Abida-Islam, P.; Muller-Schiffmann, A.; Mariappan, A.; Goureau, O.; Gruell, H.; Walker, A.; et al. SARS-CoV-2 targets neurons of 3D human brain organoids. EMBO J. 2020, 39, e106230. [Google Scholar] [CrossRef]
- Grossegesse, M.; Bourquain, D.; Neumann, M.; Schaade, L.; Schulze, J.; Mache, C.; Wolff, T.; Nitsche, A.; Doellinger, J. Deep Time Course Proteomics of SARS-CoV- and SARS-CoV-2-Infected Human Lung Epithelial Cells (Calu-3) Reveals Strong Induction of Interferon-Stimulated Gene Expression by SARS-CoV-2 in Contrast to SARS-CoV. J. Proteome Res. 2022, 21, 459–469. [Google Scholar] [CrossRef]
- Flamier, A.; Bisht, P.; Richards, A.; Tomasello, D.L.; Jaenisch, R. Human iPS cell-derived sensory neurons can be infected by SARS-CoV-2. iScience 2023, 26, 107690. [Google Scholar] [CrossRef]
- Adingupu, D.D.; Soroush, A.; Hansen, A.; Twomey, R.; Dunn, J.F. Brain hypoxia, neurocognitive impairment, and quality of life in people post-COVID-19. J. Neurol. 2023, 270, 3303–3314. [Google Scholar] [CrossRef]
- Wierzba-Bobrowicz, T.; Krajewski, P.; Tarka, S.; Acewicz, A.; Felczak, P.; Stępień, T.; Golan, M.P.; Grzegorczyk, M. Neuropathological analysis of the brains of fifty-two patients with COVID-19. Folia Neuropathol. 2021, 59, 219–231. [Google Scholar] [CrossRef]
- Kong, W.; Montano, M.; Corley, M.J.; Helmy, E.; Kobayashi, H.; Kinisu, M.; Suryawanshi, R.; Luo, X.; Royer, L.A.; Roan, N.R.; et al. Neuropilin-1 Mediates SARS-CoV-2 Infection of Astrocytes in Brain Organoids, Inducing Inflammation Leading to Dysfunction and Death of Neurons. mBio 2022, 13, e0230822. [Google Scholar] [CrossRef]
- Andrews, M.G.; Mukhtar, T.; Eze, U.C.; Simoneau, C.R.; Ross, J.; Parikshak, N.; Wang, S.; Zhou, L.; Koontz, M.; Velmeshev, D.; et al. Tropism of SARS-CoV-2 for human cortical astrocytes. Proc. Natl. Acad. Sci. USA 2022, 119, e2122236119. [Google Scholar] [CrossRef] [PubMed]
- Kase, Y.; Sonn, I.; Goto, M.; Murakami, R.; Sato, T.; Okano, H. The original strain of SARS-CoV-2, the Delta variant, and the Omicron variant infect microglia efficiently, in contrast to their inability to infect neurons: Analysis using 2D and 3D cultures. Exp. Neurol. 2023, 363, 114379. [Google Scholar] [CrossRef] [PubMed]
- Malik, J.R.; Acharya, A.; Avedissian, S.N.; Byrareddy, S.N.; Fletcher, C.V.; Podany, A.T.; Dyavar, S.R. ACE-2, TMPRSS2, and Neuropilin-1 Receptor Expression on Human Brain Astrocytes and Pericytes and SARS-CoV-2 Infection Kinetics. Int. J. Mol. Sci. 2023, 24, 8622. [Google Scholar] [CrossRef] [PubMed]
- Plantone, D.; Locci, S.; Bergantini, L.; Manco, C.; Cortese, R.; Meocci, M.; Cavallaro, D.; d’Alessandro, M.; Bargagli, E.; De Stefano, N. Brain neuronal and glial damage during acute COVID-19 infection in absence of clinical neurological manifestations. J. Neurol. Neurosurg. Psychiatry 2022, 93, 1343–1348. [Google Scholar] [CrossRef]
- Jeong, G.U.; Lyu, J.; Kim, K.D.; Chung, Y.C.; Yoon, G.Y.; Lee, S.; Hwang, I.; Shin, W.H.; Ko, J.; Lee, J.Y.; et al. SARS-CoV-2 Infection of Microglia Elicits Proinflammatory Activation and Apoptotic Cell Death. Microbiol. Spectr. 2022, 10, e0109122. [Google Scholar] [CrossRef]
- Fernandez-Castaneda, A.; Lu, P.; Geraghty, A.C.; Song, E.; Lee, M.H.; Wood, J.; O’Dea, M.R.; Dutton, S.; Shamardani, K.; Nwangwu, K.; et al. Mild respiratory COVID can cause multi-lineage neural cell and myelin dysregulation. Cell 2022, 185, 2452–2468.e16. [Google Scholar] [CrossRef]
- Samudyata Oliveira, A.O.; Malwade, S.; Rufino de Sousa, N.; Goparaju, S.K.; Gracias, J.; Orhan, F.; Steponaviciute, L.; Schalling, M.; Sheridan, S.D.; Perlis, R.H.; et al. SARS-CoV-2 promotes microglial synapse elimination in human brain organoids. Mol. Psychiatry 2022, 27, 3939–3950. [Google Scholar] [CrossRef] [PubMed]
- Stepien, T.; Tarka, S.; Chmura, N.; Grzegorczyk, M.; Acewicz, A.; Felczak, P.; Wierzba-Bobrowicz, T. Influence of SARS-CoV-2 on Adult Human Neurogenesis. Cells 2023, 12, 244. [Google Scholar] [CrossRef]
- Geyer, P.E.; Arend, F.M.; Doll, S.; Louiset, M.L.; Virreira Winter, S.; Muller-Reif, J.B.; Torun, F.M.; Weigand, M.; Eichhorn, P.; Bruegel, M.; et al. High-resolution serum proteome trajectories in COVID-19 reveal patient-specific seroconversion. EMBO Mol. Med. 2021, 13, e14167. [Google Scholar] [CrossRef]
- Messner, C.B.; Demichev, V.; Bloomfield, N.; Yu, J.S.L.; White, M.; Kreidl, M.; Egger, A.S.; Freiwald, A.; Ivosev, G.; Wasim, F.; et al. Ultra-fast proteomics with Scanning SWATH. Nat. Biotechnol. 2021, 39, 846–854. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, Z.; Zhou, J.; Han, S.; Kang, Z.; Chuang, H.; Fan, H.; Zhao, H.; Wang, L.; Ning, Y.; et al. Next-Generation Sequencing and Proteomics of Cerebrospinal Fluid From COVID-19 Patients With Neurological Manifestations. Front. Immunol. 2021, 12, 782731. [Google Scholar] [CrossRef]
- Chen, Y.C.; Pristera, A.; Ayub, M.; Swanwick, R.S.; Karu, K.; Hamada, Y.; Rice, A.S.; Okuse, K. Identification of a receptor for neuropeptide VGF and its role in neuropathic pain. J. Biol. Chem. 2013, 288, 34638–34646. [Google Scholar] [CrossRef] [PubMed]
- Thaweethai, T.; Jolley, S.E.; Karlson, E.W.; Levitan, E.B.; Levy, B.; McComsey, G.A.; McCorkell, L.; Nadkarni, G.N.; Parthasarathy, S.; Singh, U.; et al. Development of a Definition of Postacute Sequelae of SARS-CoV-2 Infection. JAMA 2023, 329, 1934–1946. [Google Scholar] [CrossRef] [PubMed]
- Reza Sailani, M.; Jahanbani, F.; Nasiri, J.; Behnam, M.; Salehi, M.; Sedghi, M.; Hoseinzadeh, M.; Takahashi, S.; Zia, A.; Gruber, J.; et al. Association of AHSG with alopecia and mental retardation (APMR) syndrome. Hum. Genet. 2017, 136, 287–296. [Google Scholar] [CrossRef]
- Geroldi, D.; Minoretti, P.; Bianchi, M.; Di Vito, C.; Reino, M.; Bertona, M.; Emanuele, E. Genetic association of alpha2-Heremans-Schmid glycoprotein polymorphism with late-onset Alzheimer’s disease in Italians. Neurosci. Lett. 2005, 386, 176–178. [Google Scholar] [CrossRef]
- Ziff, O.J.; Ashton, N.J.; Mehta, P.R.; Brown, R.; Athauda, D.; Heaney, J.; Heslegrave, A.J.; Benedet, A.L.; Blennow, K.; Checkley, A.M.; et al. Amyloid processing in COVID-19-associated neurological syndromes. J. Neurochem. 2022, 161, 146–157. [Google Scholar] [CrossRef]
- Lunnon, K.; Keohane, A.; Pidsley, R.; Newhouse, S.; Riddoch-Contreras, J.; Thubron, E.B.; Devall, M.; Soininen, H.; Kłoszewska, I.; Mecocci, P.; et al. Mitochondrial genes are altered in blood early in Alzheimer’s disease. Neurobiol. Aging 2017, 53, 36–47. [Google Scholar] [CrossRef]
- Zhou, C.; Taslima, F.; Abdelhamid, M.; Kim, S.W.; Akatsu, H.; Michikawa, M.; Jung, C.G. Beta-Amyloid Increases the Expression Levels of Tid1 Responsible for Neuronal Cell Death and Amyloid Beta Production. Mol. Neurobiol. 2020, 57, 1099–1114. [Google Scholar] [CrossRef] [PubMed]
- Pei, J.J.; Braak, H.; An, W.L.; Winblad, B.; Cowburn, R.F.; Iqbal, K.; Grundke-Iqbal, I. Up-regulation of mitogen-activated protein kinases ERK1/2 and MEK1/2 is associated with the progression of neurofibrillary degeneration in Alzheimer’s disease. Brain Res. Mol. Brain Res. 2002, 109, 45–55. [Google Scholar] [CrossRef]
- Banzhaf-Strathmann, J.; Benito, E.; May, S.; Arzberger, T.; Tahirovic, S.; Kretzschmar, H.; Fischer, A.; Edbauer, D. MicroRNA-125b induces tau hyperphosphorylation and cognitive deficits in Alzheimer’s disease. EMBO J. 2014, 33, 1667–1680. [Google Scholar] [CrossRef]
- Kristensen, A.S.; Jenkins, M.A.; Banke, T.G.; Schousboe, A.; Makino, Y.; Johnson, R.C.; Huganir, R.; Traynelis, S.F. Mechanism of Ca2+/calmodulin-dependent kinase II regulation of AMPA receptor gating. Nat. Neurosci. 2011, 14, 727–735. [Google Scholar] [CrossRef] [PubMed]
- Reshetnikov, V.V.; Kisaretova, P.E.; Ershov, N.I.; Shulyupova, A.S.; Oshchepkov, D.Y.; Klimova, N.V.; Ivanchihina, A.V.; Merkulova, T.I.; Bondar, N.P. Genes associated with cognitive performance in the Morris water maze: An RNA-seq study. Sci. Rep. 2020, 10, 22078. [Google Scholar] [CrossRef] [PubMed]
- Barbagallo, D.; Palermo, C.I.; Barbagallo, C.; Battaglia, R.; Caponnetto, A.; Spina, V.; Ragusa, M.; Di Pietro, C.; Scalia, G.; Purrello, M. Competing endogenous RNA network mediated by circ_3205 in SARS-CoV-2 infected cells. Cell Mol. Life Sci. 2022, 79, 75. [Google Scholar] [CrossRef]
- Guo, S.; Lei, X.; Chang, Y.; Zhao, J.; Wang, J.; Dong, X.; Liu, Q.; Zhang, Z.; Wang, L.; Yi, D.; et al. SARS-CoV-2 hijacks cellular kinase CDK2 to promote viral RNA synthesis. Signal Transduct. Target. Ther. 2022, 7, 400. [Google Scholar] [CrossRef]
- Nunez Lopez, Y.O.; Iliuk, A.; Casu, A.; Parikh, A.; Smith, J.S.; Corbin, K.; Lupu, D.; Pratley, R.E. Extracellular vesicle proteomics and phosphoproteomics identify pathways for increased risk in patients hospitalized with COVID-19 and type 2 diabetes mellitus. Diabetes Res. Clin. Pract. 2023, 197, 110565. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.K.; Chaubey, G.; Chen, J.Y.; Suravajhala, P. Decoding SARS-CoV-2 hijacking of host mitochondria in COVID-19 pathogenesis. Am. J. Physiol. Cell Physiol. 2020, 319, C258–C267. [Google Scholar] [CrossRef]
- Shang, C.; Liu, Z.; Zhu, Y.; Lu, J.; Ge, C.; Zhang, C.; Li, N.; Jin, N.; Li, Y.; Tian, M.; et al. SARS-CoV-2 Causes Mitochondrial Dysfunction and Mitophagy Impairment. Front. Microbiol. 2021, 12, 780768. [Google Scholar] [CrossRef]
- Ferrucci, R.; Cuffaro, L.; Capozza, A.; Rosci, C.; Maiorana, N.; Groppo, E.; Reitano, M.R.; Poletti, B.; Ticozzi, N.; Tagliabue, L.; et al. Brain positron emission tomography (PET) and cognitive abnormalities one year after COVID-19. J. Neurol. 2023, 270, 1823–1834. [Google Scholar] [CrossRef]
- Hosp, J.A.; Dressing, A.; Blazhenets, G.; Bormann, T.; Rau, A.; Schwabenland, M.; Thurow, J.; Wagner, D.; Waller, C.; Niesen, W.D.; et al. Cognitive impairment and altered cerebral glucose metabolism in the subacute stage of COVID-19. Brain 2021, 144, 1263–1276. [Google Scholar] [CrossRef]
- Thomas, T.; Stefanoni, D.; Reisz, J.A.; Nemkov, T.; Bertolone, L.; Francis, R.O.; Hudson, K.E.; Zimring, J.C.; Hansen, K.C.; Hod, E.A.; et al. COVID-19 infection alters kynurenine and fatty acid metabolism, correlating with IL-6 levels and renal status. JCI Insight 2020, 5, e140327. [Google Scholar] [CrossRef]
- Krishnan, S.; Nordqvist, H.; Ambikan, A.T.; Gupta, S.; Sperk, M.; Svensson-Akusjarvi, S.; Mikaeloff, F.; Benfeitas, R.; Saccon, E.; Ponnan, S.M.; et al. Metabolic Perturbation Associated With COVID-19 Disease Severity and SARS-CoV-2 Replication. Mol. Cell. Proteom. 2021, 20, 100159. [Google Scholar] [CrossRef] [PubMed]
- Weinhofer, I.; Buda, A.; Kunze, M.; Palfi, Z.; Traunfellner, M.; Hesse, S.; Villoria-Gonzalez, A.; Hofmann, J.; Hametner, S.; Regelsberger, G.; et al. Peroxisomal very long-chain fatty acid transport is targeted by herpesviruses and the antiviral host response. Commun. Biol. 2022, 5, 944. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, A.J.; Jahan, S.; Chaturvedi, S.; Siddiqui, M.A.; Alshahrani, M.M.; Abdelgadir, A.; Hamadou, W.S.; Saxena, J.; Sundararaj, B.K.; Snoussi, M.; et al. Therapeutic Role of ELOVL in Neurological Diseases. ACS Omega 2023, 8, 9764–9774. [Google Scholar] [CrossRef] [PubMed]
- Moolamalla, S.T.R.; Balasubramanian, R.; Chauhan, R.; Priyakumar, U.D.; Vinod, P.K. Host metabolic reprogramming in response to SARS-CoV-2 infection: A systems biology approach. Microb. Pathog. 2021, 158, 105114. [Google Scholar] [CrossRef]
- Varma, V.R.; Busra Luleci, H.; Oommen, A.M.; Varma, S.; Blackshear, C.T.; Griswold, M.E.; An, Y.; Roberts, J.A.; O’Brien, R.; Pletnikova, O.; et al. Abnormal brain cholesterol homeostasis in Alzheimer’s disease-a targeted metabolomic and transcriptomic study. NPJ Aging Mech. Dis. 2021, 7, 11. [Google Scholar] [CrossRef]
- Saher, G. Cholesterol Metabolism in Aging and Age-Related Disorders. Annu. Rev. Neurosci. 2023, 46, 59–78. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Basak, I.; Harfoot, R.; Palmer, J.E.; Kumar, A.; Quiñones-Mateu, M.E.; Schweitzer, L.; Hughes, S.M. Neuroproteomic Analysis after SARS-CoV-2 Infection Reveals Overrepresented Neurodegeneration Pathways and Disrupted Metabolic Pathways. Biomolecules 2023, 13, 1597. https://0-doi-org.brum.beds.ac.uk/10.3390/biom13111597
Basak I, Harfoot R, Palmer JE, Kumar A, Quiñones-Mateu ME, Schweitzer L, Hughes SM. Neuroproteomic Analysis after SARS-CoV-2 Infection Reveals Overrepresented Neurodegeneration Pathways and Disrupted Metabolic Pathways. Biomolecules. 2023; 13(11):1597. https://0-doi-org.brum.beds.ac.uk/10.3390/biom13111597
Chicago/Turabian StyleBasak, Indranil, Rhodri Harfoot, Jennifer E. Palmer, Abhishek Kumar, Miguel E. Quiñones-Mateu, Lucia Schweitzer, and Stephanie M. Hughes. 2023. "Neuroproteomic Analysis after SARS-CoV-2 Infection Reveals Overrepresented Neurodegeneration Pathways and Disrupted Metabolic Pathways" Biomolecules 13, no. 11: 1597. https://0-doi-org.brum.beds.ac.uk/10.3390/biom13111597