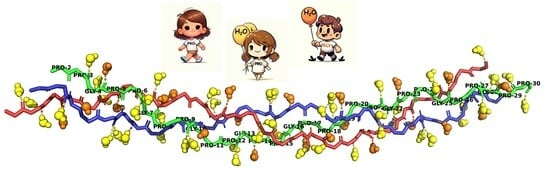
Collagen Structured Hydration
Abstract
:1. Introduction
- (a)
- The bb-carbonyl oxygen atoms of the G, X, and Y residues accepts HBs from 1, 0, and 2 WMs, respectively. Clearly, X-CO is not hydrated because it is the HB acceptor in a direct G-NH⋯OC-X HB.
- (b)
- (c)
- When an amino acid is present in the X or Y positions, or Y is Hyp, their NH or OH moieties (respectively) serve as anchor points for additional water bridges.
- (a)
- The PPG segments are short and located at the two ends of the 1BKV peptide, where the triple helix is less compact and more exposed to solution.
- (b)
- The zwitterionic peptide was not “capped” to better mimic infinitely long chains.
- (c)
- The 10 ns simulations are not sufficiently long to obtain the residence time distribution (only its average).
- (d)
- The average residence time was measured for a water H atom, rather than its O atom. This monitors water reorientation times, which terminate with a flip exchanging the two H atoms of a WM rather than the O atom leaving its binding location. Consequently, the study is best repeated for an intact (PPG) peptide.
2. Materials and Methods
3. Results
3.1. Stability and Fluctuations
3.2. Radius of Gyration
3.3. Characterization of Hydration Sites
3.3.1. Calculating Inner-Shell Hydration Numbers
- (a)
- For a single trajectory frame, or an X-ray structure PDB file, one can simply visualize the binding site and its ligands, e.g., (i) “chain A and resname PRO and name O” in one color, and (ii) “water and within 3.5 of (chain A and resname PRO and name O)” in another color. Note that for a single frame a cutoff distance of 3.5 Å is selected, which is slightly larger than the value of 3.25 Å from an ensemble average.
- (b)
- The average number of WM neighbors of a given Res-CO site can be calculated from Tcl script S1. Here, the cutoff distance, 3.225 Å, is taken from the first minimum in for liquid water (at 300 K).
- (c)
- The equilibrium water structure at the protein surface can be captured by the radial distribution function (RDF), denoted , which monitors the average number of water oxygen atoms () within a spherical shell surrounding the oxygen atom of the bb-carbonyl of each residue of the [(PPG)] peptide. The Pro(X) residue did not show any peak for the first hydration layer ( Å), because of its carbonyl oxygen HB formation with the Gly-NH of the neighboring chain. This leaves the Gly-CO and Pro(Y)-CO sites for investigation.
3.3.2. Crystal Structure First-Shell Hydration
3.3.3. Room Temperature First-Shell Hydration
3.3.4. Solvent-Accessible Surface Area (SASA)
3.3.5. Hydrogen-Bonded Second Hydration Shell
- (a)
- Use the same as above, and integrate its second peak namely, from to the second minimum, which occurs around 5.61 Å (see Figure 6). “Second neighbors” by this criterion will include WMs that are not HBed to the water in the first hydration shell (i.e., to Res-CO⋯HOH).
- (b)
- Restricting our interest to WMs that are directly HBed to first-shell waters requires that we consider another RDF, , that is centered on a first-shell WM of Res-CO. The integral of up to its first minimum, , is subsequently the number of HBed WMs in the second layer of Res-CO.
3.4. Residence Time Distribution of Water Near Res-CO
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| bb | Backbone |
| CM | Center of Mass |
| CMP | Collagen-Mimetic Peptide |
| ECM | Extracellular Matrix |
| FF | Force Field |
| HB | Hydrogen Bond |
| MD | Molecular Dynamics |
| NMR | Nuclear Magnetic Resonance |
| PDB | Protein Data Bank |
| PME | Particle Mesh Ewald |
| RDF | Radial Distribution Function |
| RMSD | Root Mean Square Deviation |
| RMSF | Root Mean Square Fluctuation |
| SASA | Solvent-Accessible Surface Area |
| VMD | Visual Molecular Dynamics |
| WM | Water Molecule |
References
- Ball, P. Water is an active matrix of life for cell and molecular biology. Proc. Nat. Acad. Sci. USA 2017, 114, 13327–13335. [Google Scholar] [CrossRef] [PubMed]
- Agmon, N. The Grotthuss mechanism. Chem. Phys. Lett. 1995, 244, 456–462. [Google Scholar] [CrossRef]
- Bellissent-Funel, M.C.; Hassanali, A.; Havenith, M.; Henchman, R.; Pohl, P.; Sterpone, F.; Van Der Spoel, D.; Xu, Y.; Garcia, A.E. Water determines the structure and dynamics of proteins. Chem. Rev. 2016, 116, 7673–7697. [Google Scholar] [CrossRef] [PubMed]
- Pentelute, B.L.; Gates, Z.P.; Tereshko, V.; Dashnau, J.L.; Vanderkooi, J.M.; Kossiakoff, A.A.; Kent, S.B. X-ray structure of snow flea antifreeze protein determined by racemic crystallization of synthetic protein enantiomers. J. Am. Chem. Soc. 2008, 130, 9695–9701. [Google Scholar] [CrossRef] [PubMed]
- Shoulders, M.D.; Raines, R.T. Collagen structure and stability. Annu. Rev. Biochem. 2009, 78, 929–958. [Google Scholar] [CrossRef]
- Bella, J. Collagen structure: New tricks from a very old dog. Biochem. J. 2016, 473, 1001–1025. [Google Scholar] [CrossRef] [PubMed]
- Rich, A.; Crick, F. The molecular structure of collagen. J. Mol. Biol. 1961, 3, 483–506. [Google Scholar] [CrossRef] [PubMed]
- Okuyama, K. Revisiting the molecular structure of collagens. Connect. Tissue Res. 2008, 49, 299–310. [Google Scholar] [CrossRef]
- Ramshaw, J.A.; Glattauer, V. The Structure of Collagen. In Biophysical and Chemical Properties of Collagen: Biomedical Applications; IOP Publishing: Bristol, UK, 2019; Chapter 2. [Google Scholar]
- Ramachandran, G.; Chandrasekharan, R. Interchain hydrogen bonds via bound water molecules in the collagen triple helix. Biopolymers 1968, 6, 1649–1658. [Google Scholar] [CrossRef]
- Bella, J.; Berman, H.M. Crystallographic evidence for Cα–H ···O=C hydrogen bonds in a collagen triple helix. J. Mol. Biol. 1996, 264, 734–742. [Google Scholar] [CrossRef]
- Fraser, R.; MacRae, T. The crystalline structure of collagen fibrils in tendon. J. Mol. Biol. 1979, 127, 129–133. [Google Scholar] [CrossRef] [PubMed]
- Okuyama, K.; Tanaka, N.; Ashida, T.; Kakudo, M.; Sakakibaka, S.; Kishida, Y. An X-ray study of the synthetic polypeptide (Pro-Pro-Gly)10. J. Mol. Biol. 1972, 72, 571–576. [Google Scholar] [CrossRef] [PubMed]
- Okuyama, K.; Okuyama, K.; Arnott, S.; Takayanagi, M.; Kakudo, M. Crystal and molecular structure of a collagen-like polypeptide (Pro-Pro-Gly)10. J. Mol. Biol. 1981, 152, 427–443. [Google Scholar] [CrossRef] [PubMed]
- Kramer, R.Z.; Vitagliano, L.; Bella, J.; Berisio, R.; Mazzarella, L.; Brodsky, B.; Zagari, A.; Berman, H.M. X-ray crystallographic determination of a collagen-like peptide with the repeating sequence (Pro-Pro-Gly). J. Mol. Biol. 1998, 280, 623–638. [Google Scholar] [CrossRef]
- Nagarajan, V.; Kamitori, S.; Okuyama, K. Crystal structure analysis of collagen model peptide (Pro-Pro-Gly)10. J. Biochem. 1998, 124, 1117–1123. [Google Scholar] [CrossRef] [PubMed]
- Berisio, R.; Vitagliano, L.; Mazzarella, L.; Zagari, A. Crystal structure of the collagen triple helix model [(Pro-Pro-Gly)10]3. Protein Sci. 2002, 11, 262–270. [Google Scholar] [CrossRef] [PubMed]
- Hongo, C.; Nagarajan, V.; Noguchi, K.; Kamitori, S.; Okuyama, K.; Tanaka, Y.; Nishino, N. Average crystal structure of (Pro-Pro-Gly)9 at 1.0 Å resolution. Polym. J. 2001, 33, 812–818. [Google Scholar] [CrossRef]
- Hongo, C.; Noguchi, K.; Okuyama, K.; Tanaka, Y.; Nishino, N. Repetitive interactions observed in the crystal structure of a collagen-model peptide, [(Pro-Pro-Gly)9]3. J. Biochem. 2005, 138, 135–144. [Google Scholar] [CrossRef]
- Berendsen, H. Nuclear magnetic resonance study of collagen hydration. J. Chem. Phys. 1962, 36, 3297–3305. [Google Scholar] [CrossRef]
- Bella, J.; Eaton, M.; Brodsky, B.; Berman, H.M. Crystal and molecular structure of a collagen-like peptide at 1.9 Å resolution. Science 1994, 266, 75–81. [Google Scholar] [CrossRef]
- Bella, J.; Brodsky, B.; Berman, H.M. Hydration structure of a collagen peptide. Structure 1995, 3, 893–906. [Google Scholar] [CrossRef] [PubMed]
- Brodsky, B.; Ramshaw, J.A. The collagen triple-helix structure. Matrix Biol. 1997, 15, 545–554. [Google Scholar] [CrossRef] [PubMed]
- Kramer, R.Z.; Berman, H.M. Patterns of hydration in crystalline collagen peptides. J. Biomol. Struct. Dyn. 1998, 16, 367–380. [Google Scholar] [CrossRef] [PubMed]
- Fullerton, G.D.; Rahal, A. Collagen structure: The molecular source of the tendon magic angle effect. J. Magn. Reson. Imaging 2007, 25, 345–361. [Google Scholar] [CrossRef] [PubMed]
- Fullerton, G.; Cameron, I.; Ord, V. Orientation of tendons in the magnetic field and its effect on T2 relaxation times. Radiology 1985, 155, 433–435. [Google Scholar] [CrossRef] [PubMed]
- Holmgren, S.K.; Taylor, K.M.; Bretscher, L.E.; Raines, R.T. Code for collagen’s stability deciphered. Nature 1998, 392, 666–667. [Google Scholar] [CrossRef] [PubMed]
- Klein, T.E.; Huang, C.C. Computational investigations of structural changes resulting from point mutations in a collagen-like peptide. Biopolymers 1999, 49, 167–183. [Google Scholar] [CrossRef]
- Mogilner, I.G.; Ruderman, G.; Grigera, J.R. Collagen stability, hydration and native state. J. Mol. Graph. Model. 2002, 21, 209–213. [Google Scholar] [CrossRef]
- De Simone, A.; Vitagliano, L.; Berisio, R. Role of hydration in collagen triple helix stabilization. Biochem. Biophys. Res. Commun. 2008, 372, 121–125. [Google Scholar] [CrossRef]
- Ravikumar, K.M.; Hwang, W. Region-specific role of water in collagen unwinding and assembly. Proteins: Struct. Funct. Bioinform. 2008, 72, 1320–1332. [Google Scholar] [CrossRef]
- Gough, C.A.; Anderson, R.W.; Bhatnagar, R.S. The role of bound water in the stability of the triple-helical conformation of (Pro-Pro-Gly)10. J. Biomol. Struct. Dyn. 1998, 15, 1029–1037. [Google Scholar] [CrossRef]
- Tourell, M.C.; Momot, K.I. Molecular dynamics of a hydrated collagen peptide: Insights into rotational motion and residence times of single-water bridges in collagen. J. Phys. Chem. B 2016, 120, 12432–12443. [Google Scholar] [CrossRef] [PubMed]
- Madhavi, W.M.; Weerasinghe, S.; Fullerton, G.D.; Momot, K.I. Structure and dynamics of collagen hydration water from molecular dynamics simulations: Implications of temperature and pressure. J. Phys. Chem. B 2019, 123, 4901–4914. [Google Scholar] [CrossRef] [PubMed]
- Kramer, R.Z.; Bella, J.; Mayville, P.; Brodsky, B.; Berman, H.M. Sequence dependent conformational variations of collagen triple-helical structure. Nat. Struct. Mol. Biol. 1999, 6, 454–457. [Google Scholar]
- Bryan, M.A.; Brauner, J.W.; Anderle, G.; Flach, C.R.; Brodsky, B.; Mendelsohn, R. FTIR studies of collagen model peptides: Complementary experimental and simulation approaches to conformation and unfolding. J. Am. Chem. Soc. 2007, 129, 7877–7884. [Google Scholar] [CrossRef] [PubMed]
- Wu, D. Proline puckering parameters for collagen structure simulations. AIP Adv. 2015, 5, 037124. [Google Scholar] [CrossRef]
- Salomon-Ferrer, R.; Case, D.A.; Walker, R.C. An overview of the Amber biomolecular simulation package. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2013, 3, 198–210. [Google Scholar] [CrossRef]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef]
- Huang, J.; Rauscher, S.; Nawrocki, G.; Ran, T.; Feig, M.; De Groot, B.L.; Grubmüller, H.; MacKerell, A.D. CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nat. Methods 2017, 14, 71–73. [Google Scholar] [CrossRef]
- Abascal, J.L.; Vega, C. A general purpose model for the condensed phases of water: TIP4P/2005. J. Chem. Phys. 2005, 123, 234505. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef]
- Van Gunsteren, W.; Berendsen, H.; Hermans, J.; Hol, W.; Postma, J. Computer simulation of the dynamics of hydrated protein crystals and its comparison with X-ray data. Proc. Nat. Acad. Sci. USA 1983, 80, 4315–4319. [Google Scholar] [CrossRef] [PubMed]
- Wall, M.E.; Calabró, G.; Bayly, C.I.; Mobley, D.L.; Warren, G.L. Biomolecular solvation structure revealed by molecular dynamics simulations. J. Am. Chem. Soc. 2019, 141, 4711–4720. [Google Scholar] [CrossRef] [PubMed]
- Caldararu, O.; Ignjatović, M.M.; Oksanen, E.; Ryde, U. Water structure in solution and crystal molecular dynamics simulations compared to protein crystal structures. RSC Adv. 2020, 10, 8435–8443. [Google Scholar] [CrossRef] [PubMed]
- Streeter, I.; de Leeuw, N.H. Atomistic modeling of collagen proteins in their fibrillar environment. J. Phys. Chem. B 2010, 114, 13263–13270. [Google Scholar] [CrossRef] [PubMed]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An Nlog (N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Parrinello, M.; Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Messias, A.; Santos, D.; Pontes, F.; Lima, F.; Soares, T. Out of sight, out of mind: The effect of the equilibration protocol on the structural ensembles of charged glycolipid bilayers. Molecules 2020, 25, 5120. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.; Fraaije, J.G. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Tarannum, A.; Muvva, C.; Mehta, A.; Rao, J.R.; Fathima, N.N. Phosphonium based ionic liquids-stabilizing or destabilizing agents for collagen? RSC Adv. 2016, 6, 4022–4033. [Google Scholar] [CrossRef]
- Zimm, B.H.; Stockmayer, W.H. The dimensions of chain molecules containing branches and rings. J. Chem. Phys. 1949, 17, 1301–1314. [Google Scholar] [CrossRef]
- Flory, P.J.; Volkenstein, M. Statistical Mechanics of Chain Molecules; Wiley Online Library: Hoboken, NJ, USA, 1969. [Google Scholar]
- Walker, K.T.; Nan, R.; Wright, D.W.; Gor, J.; Bishop, A.C.; Makhatadze, G.I.; Brodsky, B.; Perkins, S.J. Non-linearity of the collagen triple helix in solution and implications for collagen function. Biochem. J. 2017, 474, 2203–2217. [Google Scholar] [CrossRef] [PubMed]
- Skinner, L.B.; Huang, C.; Schlesinger, D.; Pettersson, L.G.; Nilsson, A.; Benmore, C.J. Benchmark oxygen-oxygen pair-distribution function of ambient water from X-ray diffraction measurements with a wide Q-range. J. Chem. Phys. 2013, 138, 074506. [Google Scholar] [CrossRef] [PubMed]
- McPherson, A., Jr. Crystallization of proteins from polyethylene glycol. J. Biol. Chem. 1976, 251, 6300–6303. [Google Scholar] [CrossRef] [PubMed]
- Shrake, A.; Rupley, J.A. Environment and exposure to solvent of protein atoms. Lysozyme and insulin. J. Mol. Biol. 1973, 79, 351–371. [Google Scholar] [CrossRef]
- Bryant, R.G. The NMR Time Scale. J. Chem. Educ. 1983, 60, 933–935. [Google Scholar] [CrossRef]
- Arkhipov, V.I.; Agmon, N. Relation between macroscopic and microscopic dielectric relaxation times in water dynamics. Isr. J. Chem. 2003, 43, 363–371. [Google Scholar] [CrossRef]
- Narang, P.; Bhushan, K.; Bose, S.; Jayaram, B. A computational pathway for bracketing native-like structures for small alpha helical globular proteins. Phys. Chem. Chem. Phys. 2005, 7, 2364–2375. [Google Scholar] [CrossRef]
- Smilgies, D.; Folta-Stogniew, E. Molecular weight–gyration radius relation of globular proteins: A comparison of light scattering, small-angle X-ray scattering and structure-based data. J. Appl. Cryst. 2015, 48, 1604–1606. [Google Scholar] [CrossRef]














| Temperature | |||||
|---|---|---|---|---|---|
| 300 K | 11.95 | 14.49 | 18.9 | 24.0 | 25.6 |
| 250 K | 11.91 | 14.36 | 18.96 | 23.9 | 25.5 |
| 100 K (X-ray) | 23.51 | 25.37 |
| 32.78 | 38.84 | 54.40 | 72.56 (70.66) | 81.41 (80.03) | |
| 0.3645 | 0.3731 | 0.347 | 0.3308 (0.3382) | 0.3145 (0.3186) |
| Temperature | Water/Gly-CO | Water/Pro(Y)-CO |
|---|---|---|
| 300 K | 1.041 | 1.61 |
| 250 K | 1.056 | 1.70 |
| Temperature | Gly-CO (Å) | Pro(Y)-CO (Å) |
|---|---|---|
| 300 K | 6.43 | 15.03 |
| 250 K | 6.56 | 15.85 |
| Temperature | Gly (Å) | Pro(X) (Å) | Pro(Y) (Å) |
|---|---|---|---|
| 300 K | 8.45 | 74.71 | 96.09 |
| 250 K | 10.57 | 78.79 | 101.87 |
| Temperature | Water/Gly-CO | Water/Pro(Y)-CO |
|---|---|---|
| 300 K | 2.26 | 2.93 |
| 250 K | 2.37 | 3.01 |
| System | A | (ps) | |
|---|---|---|---|
| Gly-CO (300 K) | 129.23 | 81.72 | 0.98 |
| Gly-CO (250 K) | 5.88 | 129.17 | 0.69 |
| Pro(Y)-CO (300 K) | 1258.0 | 25.68 | 0.99 |
| Pro(Y)-CO (250 K) | 590.81 | 39.34 | 0.98 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Biswal, S.; Agmon, N. Collagen Structured Hydration. Biomolecules 2023, 13, 1744. https://0-doi-org.brum.beds.ac.uk/10.3390/biom13121744
Biswal S, Agmon N. Collagen Structured Hydration. Biomolecules. 2023; 13(12):1744. https://0-doi-org.brum.beds.ac.uk/10.3390/biom13121744
Chicago/Turabian StyleBiswal, Satyaranjan, and Noam Agmon. 2023. "Collagen Structured Hydration" Biomolecules 13, no. 12: 1744. https://0-doi-org.brum.beds.ac.uk/10.3390/biom13121744