Self-Control of Inflammation during Tail Regeneration of Lizards
Abstract
:1. Introduction
2. Rapid Wound Healing and Protection of Antibacterial Peptides
3. Infiltration of Leukocytes to the Wounded Tail
4. Self-Blocking of the Proinflammatory Signal Pathway in Leukocytes
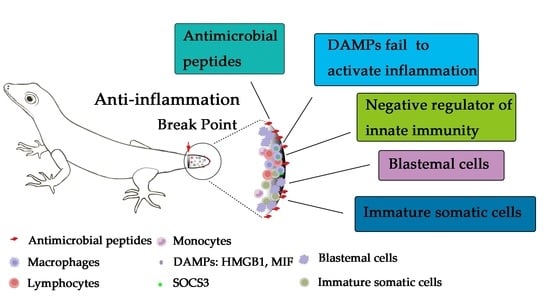
5. Crosstalk between Inflammation and Blastemal Cells or Immature Somatic Cells
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Whitby, D.J.; Ferguson, M.W. Immunohistochemical localization of growth factors in fetal wound healing. Dev. Biol. 1991, 147, 207–215. [Google Scholar] [CrossRef]
- Ferguson, M.W.; O’Kane, S. Scar-free healing: From embryonic mechanisms to adult therapeutic intervention. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2004, 359, 839–850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernando, W.A.; Leininger, E.; Simkin, J.; Li, N.; Malcom, C.A.; Sathyamoorthi, S.; Han, M.; Muneoka, K. Wound healing and blastema formation in regenerating digit tips of adult mice. Dev. Biol. 2011, 350, 301–310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, M.; Yang, X.; Lee, J.; Allan, C.H.; Muneoka, K. Development and regeneration of the neonatal digit tip in mice. Dev. Biol. 2008, 315, 125–135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seifert, A.W.; Kiama, S.G.; Seifert, M.G.; Goheen, J.R.; Palmer, T.M.; Maden, M. Skin shedding and tissue regeneration in African spiny mice (Acomys). Nature 2012, 489, 561–565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilbert, E.A.; Payne, S.L.; Vickaryous, M.K. The anatomy and histology of caudal autotomy and regeneration in lizards. Physiol. Biochem. Zool. 2013, 86, 631–644. [Google Scholar] [CrossRef] [PubMed]
- Peacock, H.M.; Gilbert, E.A.; Vickaryous, M.K. Scar-free cutaneous wound healing in the leopard gecko, Eublepharis macularius. J. Anat. 2015, 227, 596–610. [Google Scholar] [CrossRef] [Green Version]
- Khyeam, S.; Lee, S.; Huang, G.N. Genetic, Epigenetic, and Post-Transcriptional Basis of Divergent Tissue Regenerative Capacities among Vertebrates. Adv. Genet. 2021, 2, e10042. [Google Scholar]
- Daponte, V.; Tylzanowski, P.; Forlino, A. Appendage Regeneration in Vertebrates: What Makes This Possible? Cells 2021, 10, 242. [Google Scholar] [CrossRef]
- Delorme, S.L.; Lungu, I.M.; Vickaryous, M.K. Scar-free wound healing and regeneration following tail loss in the leopard gecko, Eublepharis macularius. Anat. Rec. 2012, 295, 1575–1595. [Google Scholar] [CrossRef]
- Alibardi, L. Review: Biological and Molecular Differences between Tail Regeneration and Limb Scarring in Lizard: An Inspiring Model Addressing Limb Regeneration in Amniotes. J. Exp. Zool. B Mol. Dev. Evol. 2017, 328, 493–514. [Google Scholar] [CrossRef]
- Alibardi, L. Ultrastructural features of the process of wound healing after Tail and limb amputation in lizard. Acta Zool. 2010, 91, 306–318. [Google Scholar] [CrossRef]
- King, B.L.; Yin, V.P. A Conserved MicroRNA Regulatory Circuit Is Differentially Controlled during Limb/Appendage Regeneration. PLoS ONE 2016, 11, e0157106. [Google Scholar] [CrossRef] [Green Version]
- Singh, B.N.; Koyano-Nakagawa, N.; Donaldson, A.; Weaver, C.V.; Garry, M.G.; Garry, D.J. Hedgehog Signaling during Appendage Development and Regeneration. Genes 2015, 6, 417–435. [Google Scholar] [CrossRef] [Green Version]
- McLean, K.E.; Vickaryous, M.K. A novel amniote model of epimorphic regeneration: The leopard gecko, Eublepharis macularius. BMC Dev. Biol. 2011, 11, 50. [Google Scholar] [CrossRef] [Green Version]
- Jacyniak, K.; McDonald, R.P.; Vickaryous, M.K. Tail regeneration and other phenomena of wound healing and tissue restoration in lizards. J. Exp. Biol. 2017, 220 Pt 16, 2858–2869. [Google Scholar] [CrossRef] [Green Version]
- Alibardi, L. Morphological and cellular aspects of tail and limb regeneration in lizards. A model system with implications for tissue regeneration in mammals. Adv. Anat. Embryol. Cell Biol. 2010, 207, 1–109. [Google Scholar]
- Oguro-Okano, M.; Honda, M.; Yamazaki, K.; Okano, K. Mutations in the melanocortin 1 receptor, beta-defensin103 and agouti signaling protein genes, and their association with coat color phenotypes in Akita-inu dogs. J. Vet. Med. Sci. 2011, 73, 853–858. [Google Scholar] [CrossRef] [Green Version]
- van Hoek, M.L. Antimicrobial peptides in reptiles. Pharmaceuticals 2014, 7, 723–753. [Google Scholar] [CrossRef] [Green Version]
- Lehrer, R.I.; Lu, W. Alpha-Defensins in human innate immunity. Immunol. Rev. 2012, 245, 84–112. [Google Scholar] [CrossRef]
- Zhao, L.; Lu, W. Defensins in innate immunity. Curr. Opin. Hematol. 2014, 21, 37–42. [Google Scholar] [CrossRef]
- Dalla Valle, L.; Benato, F.; Maistro, S.; Quinzani, S.; Alibardi, L. Bioinformatic and molecular characterization of beta-defensins-like peptides isolated from the green lizard Anolis carolinensis. Dev. Comp. Immunol. 2012, 36, 222–229. [Google Scholar] [CrossRef]
- Alibardi, L.; Celeghin, A.; Dalla Valle, L. Wounding in lizards results in the release of beta-defensins at the wound site and formation of an antimicrobial barrier. Dev. Comp. Immunol. 2012, 36, 557–565. [Google Scholar] [CrossRef]
- Alibardi, L. Granulocytes of reptilian sauropsids contain beta-defensin-like peptides: A comparative ultrastructural survey. J. Morphol. 2013, 274, 877–886. [Google Scholar] [CrossRef]
- Cole, A.M.; Shi, J.; Ceccarelli, A.; Kim, Y.H.; Park, A.; Ganz, T. Inhibition of neutrophil elastase prevents cathelicidin activation and impairs clearance of bacteria from wounds. Blood 2001, 97, 297–304. [Google Scholar] [CrossRef] [Green Version]
- Tongaonkar, P.; Golji, A.E.; Tran, P.; Ouellette, A.J.; Selsted, M.E. High fidelity processing and activation of the human alpha-defensin HNP1 precursor by neutrophil elastase and proteinase 3. PLoS ONE 2012, 7, e32469. [Google Scholar] [CrossRef] [Green Version]
- Ponkham, P.; Daduang, S.; Kitimasak, W.; Krittanai, C.; Chokchaichamnankit, D.; Srisomsap, C.; Svasti, J.; Kawamura, S.; Araki, T.; Thammasirirak, S. Complete amino acid sequence of three reptile lysozymes. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2010, 151, 75–83. [Google Scholar] [CrossRef]
- Kosciuczuk, E.M.; Lisowski, P.; Jarczak, J.; Strzalkowska, N.; Jozwik, A.; Horbanczuk, J.; Krzyzewski, J.; Zwierzchowski, L.; Bagnicka, E. Cathelicidins: Family of antimicrobial peptides. A review. Mol. Biol. Rep. 2012, 39, 10957–10970. [Google Scholar] [CrossRef] [Green Version]
- Dalla Valle, L.; Benato, F.; Paccanaro, M.C.; Alibardi, L. Bioinformatic and molecular characterization of cathelicidin-like peptides isolated from the green lizard Anolis carolinensis. Ital. J. Zool. 2013, 80, 177–186. [Google Scholar] [CrossRef]
- Alibardi, L. Ultrastructural immunolocalization of chatelicidin-like peptides in granulocytes of normal and regenerating lizard tissues. Acta Histochem. 2014, 116, 363–371. [Google Scholar] [CrossRef]
- Alibardi, L. Immunocytochemical detection of beta-defensins and cathelicidins in the secretory granules of the tongue in the lizard Anolis carolinensis. Acta Histochem. 2015, 117, 223–227. [Google Scholar] [CrossRef] [PubMed]
- Holthaus, K.B.; Spisni, E.; Alibardi, L. Microbicide Activity of Two Reptilian Antimicrobial Peptides on Gram Positive and Gram Negative Bacteria. J. Immun. Biol. 2016, 1, 104. [Google Scholar]
- Alibardi, L. Ultrastructural immunolocalization of antimicrobial peptides targeting bacteria in the corneous layer supports the presence of an antimicrobial barrier in reptilian epidermis. J. Cytol. Hisitol. 2016, 1, 1–7. [Google Scholar]
- Reinke, J.M.; Sorg, H. Wound repair and regeneration. Eur. Surg. Res. 2012, 49, 35–43. [Google Scholar] [CrossRef]
- Singer, A.J.; Clark, R.A. Cutaneous wound healing. N. Engl. J. Med. 1999, 341, 738–746. [Google Scholar] [CrossRef]
- Bielefeld, K.A.; Amini-Nik, S.; Alman, B.A. Cutaneous wound healing: Recruiting developmental pathways for regeneration. Cell. Mol. Life Sci. 2013, 70, 2059–2081. [Google Scholar] [CrossRef] [Green Version]
- Schultz, G.S.; Davidson, J.M.; Kirsner, R.S.; Bornstein, P.; Herman, I.M. Dynamic reciprocity in the wound microenvironment. Wound Repair Regen. 2011, 19, 134–148. [Google Scholar] [CrossRef] [Green Version]
- Gadani, S.P.; Walsh, J.T.; Lukens, J.R.; Kipnis, J. Dealing with Danger in the CNS: The Response of the Immune System to Injury. Neuron 2015, 87, 47–62. [Google Scholar] [CrossRef] [Green Version]
- Mescher, A.L.; Neff, A.W. Limb regeneration in amphibians: Immunological considerations. Sci. World J. 2006, 6 (Suppl. S1), 1–11. [Google Scholar] [CrossRef]
- Delavary, B.M.; van der Veer, W.M.; van Egmond, M.; Niessen, F.B.; Beelen, R.H. Macrophages in skin injury and repair. Immunobiology 2011, 216, 753–762. [Google Scholar] [CrossRef]
- Canfield, P.J. Comparative cell morphology in the peripheral blood film from exotic and native animals. Aust. Vet. J. 1998, 76, 793–800. [Google Scholar] [CrossRef]
- Park, J.E.; Barbul, A. Understanding the role of immune regulation in wound healing. Am. J. Surg. 2004, 187, 11S–16S. [Google Scholar] [CrossRef]
- Wang, Y.; Wei, S.; Song, H.; Zhang, X.; Wang, W.; Du, N.; Song, T.; Liang, H.; Chen, X.; Wang, Y. Macrophage migration inhibitory factor derived from spinal cord is involved in activation of macrophages following gecko tail amputation. FASEB J. 2019, 33, 14798–14810. [Google Scholar] [CrossRef] [Green Version]
- Godwin, J.W.; Pinto, A.R.; Rosenthal, N.A. Macrophages are required for adult salamander limb regeneration. Proc. Natl. Acad. Sci. USA 2013, 110, 9415–9420. [Google Scholar] [CrossRef] [Green Version]
- Simkin, J.; Gawriluk, T.R.; Gensel, J.C.; Seifert, A.W. Macrophages are necessary for epimorphic regeneration in African spiny mice. eLife 2017, 6, e24623. [Google Scholar] [CrossRef]
- Alibardi, L. Tail regeneration reduction in lizards after repetitive amputation or cauterization reflects an increase of immune cells in blastemas. Zool. Res. 2018, 39, 413–423. [Google Scholar] [CrossRef]
- Alibardi, L. Immunolocalization of 5BrdU long retaining labeled cells and macrophage infiltration in the scarring limb of lizard after limb amputation. Tissue Cell 2016, 48, 197–207. [Google Scholar] [CrossRef]
- Vitulo, N.; Dalla Valle, L.; Skobo, T.; Valle, G.; Alibardi, L. Transcriptome analysis of the regenerating tail vs. the scarring limb in lizard reveals pathways leading to successful vs. unsuccessful organ regeneration in amniotes. Dev. Dyn. 2017, 246, 116–134. [Google Scholar] [CrossRef] [Green Version]
- Brancato, S.K.; Albina, J.E. Wound macrophages as key regulators of repair: Origin, phenotype, and function. Am. J. Pathol. 2011, 178, 19–25. [Google Scholar] [CrossRef]
- Alibardi, L. Autoradiography and inmmunolabeling suggests that lizard blastema contains arginase-positive M2-like macrophages that may support tail regeneration. Ann. Anat. 2020, 231, 151549. [Google Scholar] [CrossRef]
- Alibardi, L. Immunoreactivity for Dab2 and Foxp3 suggests that immune-suppressive cells are present in the regenerating tail blastema of lizard. Acta Zool. 2021. [Google Scholar] [CrossRef]
- Londono, R.; Tighe, S.; Milnes, B.; DeMoya, C.; Quijano, L.M.; Hudnall, M.L.; Nguyen, J.; Tran, E.; Badylak, S.; Lozito, T.P. Single Cell Sequencing Analysis of Lizard Phagocytic Cell Populations and Their Role in Tail Regeneration. J. Immunol. Regen. Med. 2020, 8, 100029. [Google Scholar] [CrossRef]
- Rowley, A.F. The evolution of inflammatory mediators. Mediat. Inflamm. 1996, 5, 3–13. [Google Scholar] [CrossRef]
- Akira, S.; Uematsu, S.; Takeuchi, O. Pathogen recognition and innate immunity. Cell 2006, 124, 783–801. [Google Scholar] [CrossRef] [Green Version]
- Mandujano-Tinoco, E.A.; Sultan, E.; Ottolenghi, A.; Gershoni-Yahalom, O.; Rosental, B. Evolution of Cellular Immunity Effector Cells; Perspective on Cytotoxic and Phagocytic Cellular Lineages. Cells 2021, 10, 1853. [Google Scholar] [CrossRef]
- Seok, J.K.; Kang, H.C.; Cho, Y.Y.; Lee, H.S.; Lee, J.Y. Therapeutic regulation of the NLRP3 inflammasome in chronic inflammatory diseases. Arch. Pharm. Res. 2021, 44, 16–35. [Google Scholar] [CrossRef]
- Priyam, M.; Tripathy, M.; Rai, U.; Ghorai, S.M. Tracing the evolutionary lineage of pattern recognition receptor homologues in vertebrates: An insight into reptilian immunity via de novo sequencing of the wall lizard splenic transcriptome. Vet. Immunol. Immunopathol. 2016, 172, 26–37. [Google Scholar] [CrossRef]
- Vitulo, N.; Dalla Valle, L.; Skobo, T.; Valle, G.; Alibardi, L. Downregulation of lizard immuno-genes in the regenerating tail and myogenes in the scarring limb suggests that tail regeneration occurs in an immuno-privileged organ. Protoplasma 2017, 254, 2127–2141. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; He, B.; Li, H.; Wang, Y.; Zhou, Y.; Wang, W.; Song, T.; Du, N.; Gu, X.; Luo, Y.; et al. SOCS3 Attenuates GM-CSF/IFN-gamma-Mediated Inflammation During Spontaneous Spinal Cord Regeneration. Neurosci. Bull. 2020, 36, 778–792. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Gu, Y.; Huan, Y.; Wang, Y.; Liu, Y.; Liu, M.; Ding, F.; Gu, X.; Wang, Y. HMGB1 protein does not mediate the inflammatory response in spontaneous spinal cord regeneration: A hint for CNS regeneration. J. Biol. Chem. 2013, 288, 18204–18218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tokuyama, M.A.; Xu, C.; Fisher, R.E.; Wilson-Rawls, J.; Kusumi, K.; Newbern, J.M. Developmental and adult-specific processes contribute to de novo neuromuscular regeneration in the lizard tail. Dev. Biol. 2018, 433, 287–296. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhou, Y.; Chen, S.; Hu, Y.; Zhu, Z.; Wang, Y.; Du, N.; Song, T.; Yang, Y.; Guo, A.; et al. Macrophage migration inhibitory factor facilitates prostaglandin E2 production of astrocytes to tune inflammatory milieu following spinal cord injury. J. Neuroinflamm. 2019, 16, 85. [Google Scholar] [CrossRef]
- Su, Y.; Wang, Y.; Zhou, Y.; Zhu, Z.; Zhang, Q.; Zhang, X.; Wang, W.; Gu, X.; Guo, A.; Wang, Y. Macrophage migration inhibitory factor activates inflammatory responses of astrocytes through interaction with CD74 receptor. Oncotarget 2017, 8, 2719–2730. [Google Scholar] [CrossRef] [Green Version]
- Du, N.; Li, H.; Sun, C.; He, B.; Yang, T.; Song, H.; Wang, Y.; Wang, Y. Adult astrocytes from reptiles are resistant to proinflammatory activation via sustaining Vav1 expression. J. Biol. Chem. 2021, 296, 100527. [Google Scholar] [CrossRef]
- Khaire, K.; Verma, U.; Buch, P.; Patel, S.; Ranadive, I.; Balakrishnan, S. Site-specific variation in the activity of COX-2 alters the pattern of wound healing in the tail and limb of northern house gecko by differentially regulating the expression of local inflammatory mediators. Zoology 2021, 148, 125947. [Google Scholar] [CrossRef]
- Tanaka, E.M.; Reddien, P.W. The cellular basis for animal regeneration. Dev. Cell 2011, 21, 172–185. [Google Scholar] [CrossRef] [Green Version]
- Londono, R.; Wenzhong, W.; Wang, B.; Tuan, R.S.; Lozito, T.P. Cartilage and Muscle Cell Fate and Origins during Lizard Tail Regeneration. Front. Bioeng. Biotechnol. 2017, 5, 70. [Google Scholar] [CrossRef] [Green Version]
- Iribarne, M. Inflammation induces zebrafish regeneration. Neural. Regen. Res. 2021, 16, 1693–1701. [Google Scholar] [CrossRef]
- Hasegawa, T.; Hall, C.J.; Crosier, P.S.; Abe, G.; Kawakami, K.; Kudo, A.; Kawakami, A. Transient inflammatory response mediated by interleukin-1beta is required for proper regeneration in zebrafish fin fold. eLife 2017, 6, e22716. [Google Scholar] [CrossRef]
- Mastellos, D.C.; Deangelis, R.A.; Lambris, J.D. Complement-triggered pathways orchestrate regenerative responses throughout phylogenesis. Semin. Immunol. 2013, 25, 29–38. [Google Scholar] [CrossRef] [Green Version]
- Alibardi, L. Hyaluronic acid in the tail and limb of amphibians and lizards recreates permissive embryonic conditions for regeneration due to its hygroscopic and immunosuppressive properties. J. Exp. Zool. B Mol. Dev. Evol. 2017, 328, 760–771. [Google Scholar] [CrossRef]
- Bosak, V.; Murata, K.; Bludau, O.; Brand, M. Role of the immune response in initiating central nervous system regeneration in vertebrates: Learning from the fish. Int. J. Dev. Biol. 2018, 62, 403–417. [Google Scholar] [CrossRef] [Green Version]
- Yang, D.; de Haan, G. Inflammation and Aging of Hematopoietic Stem Cells in Their Niche. Cells 2021, 10, 1849. [Google Scholar] [CrossRef]
- Lenero, C.; Bowles, A.C.; Correa, D.; Kouroupis, D. Characterization and response to inflammatory stimulation of human endometrial-derived mesenchymal stem/stromal cells. Cytotherapy 2021, in press. [Google Scholar] [CrossRef]
- Murawala, H.; Ranadive, I.; Patel, S.; Desai, I.; Balakrishnan, S. Protein expression pattern and analysis of differentially expressed peptides during various stages of tail regeneration in Hemidactylus flaviviridis. Mech. Dev. 2018, 150, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhou, Q.; Wang, Y.; Luo, L.; Yang, J.; Yang, L.; Liu, M.; Li, Y.; Qian, T.; Zheng, Y.; et al. Gekko japonicus genome reveals evolution of adhesive toe pads and tail regeneration. Nat. Commun. 2015, 6, 10033. [Google Scholar] [CrossRef] [PubMed]
- Ge, X.; Shi, K.; Hou, J.; Fu, Y.; Xiao, H.; Chi, F.; Xu, J.; Cai, F.; Bai, C. Galectin-1 secreted by bone marrow-derived mesenchymal stem cells mediates anti-inflammatory responses in acute airway disease. Exp. Cell Res. 2021, 407, 112788. [Google Scholar] [CrossRef]
- Wang, L.; Li, Y.; Xu, M.; Deng, Z.; Zhao, Y.; Yang, M.; Liu, Y.; Yuan, R.; Sun, Y.; Zhang, H.; et al. Regulation of Inflammatory Cytokine Storms by Mesenchymal Stem Cells. Front. Immunol. 2021, 12, 726909. [Google Scholar] [CrossRef]
- Yu, S.H.; Kim, S.; Kim, Y.; Lee, S.E.; Park, J.H.; Cho, G.; Ha, J.C.; Jung, H.; Lim, S.M.; Han, K.; et al. Human umbilical cord mesenchymal stem cell-derived mitochondria (PN-101) attenuate LPS-induced inflammatory responses by inhibiting NFkappaB signaling pathway. BMB Rep. 2021, in press. [Google Scholar]
- Kokaia, Z.; Martino, G.; Schwartz, M.; Lindvall, O. Cross-talk between neural stem cells and immune cells: The key to better brain repair? Nat. Neurosci. 2012, 15, 1078–1087. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, B.; Song, H.; Wang, Y. Self-Control of Inflammation during Tail Regeneration of Lizards. J. Dev. Biol. 2021, 9, 48. https://0-doi-org.brum.beds.ac.uk/10.3390/jdb9040048
He B, Song H, Wang Y. Self-Control of Inflammation during Tail Regeneration of Lizards. Journal of Developmental Biology. 2021; 9(4):48. https://0-doi-org.brum.beds.ac.uk/10.3390/jdb9040048
Chicago/Turabian StyleHe, Bingqiang, Honghua Song, and Yongjun Wang. 2021. "Self-Control of Inflammation during Tail Regeneration of Lizards" Journal of Developmental Biology 9, no. 4: 48. https://0-doi-org.brum.beds.ac.uk/10.3390/jdb9040048