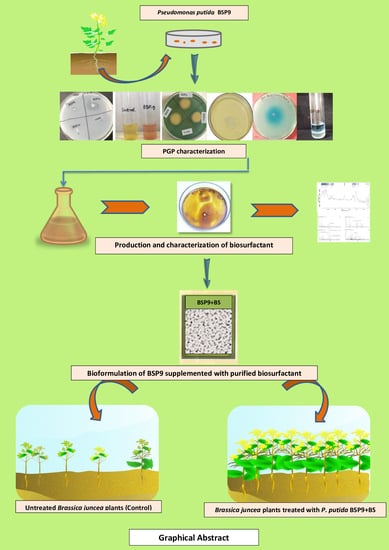
Novel Bioformulations Developed from Pseudomonas putida BSP9 and Its Biosurfactant for Growth Promotion of Brassica juncea (L.)
Abstract
:1. Introduction
2. Results and Discussion
2.1. Isolation and Identification of Isolate
2.2. PGP Characterization of the Isolate
2.3. Production, Extraction and Purification of Biosurfactant by Isolate BSP9
2.4. Structural Characterization of the Biosurfactant
2.5. Field Trial
3. Materials and Methods
3.1. Isolation and Identification of the Isolates
3.2. Plant Growth Promoting Characters of the Isolate BSP9
3.3. Production, Extraction and Purification of Biosurfactant by Isolate BSP9
3.4. Structural Characterization of the Biosurfactant
3.5. Field Study
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- How to Feed the World in 2050. Available online: http://www.fao.org/fileadmin/templates/wsfs/docs/expert_paper/How_to_Feed_the_World_in_2050.pdf (accessed on 1 August 2020).
- Arora, N.K.; Fatima, T.; Mishra, I.; Verma, M.; Mishra, J.; Mishra, V. Environmental sustainability: Challenges and viable solutions. Environ. Sustain. 2018, 1, 309–340. [Google Scholar] [CrossRef]
- Arora, N.K.; Mishra, I. United Nations sustainable development goals 2030 and environmental sustainability: Race against time. Environ. Sustain. 2019, 2, 339–342. [Google Scholar] [CrossRef] [Green Version]
- Backer, R.; Rokem, J.S.; Ilangumaran, G.; Lamont, J.; Praslickova, D.; Ricci, E.; Subramanian, S.; Smith, D.L. Plant growth-promoting rhizobacteria: Context, mechanisms of action, and roadmap to commercialization of biostimulants for sustainable agriculture. Front. Plant. Sci. 2018, 9, 1473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banat, I.M.; Franzetti, A.; Gandolfi, I.; Bestetti, G.; Martinotti, M.G.; Fracchia, L.; Smyth, T.J.; Marchant, R. Microbial biosurfactants production, applications and future potential. Appl. Microbiol. Biotechnol. 2010, 87, 427–444. [Google Scholar] [CrossRef] [PubMed]
- Egamberdieva, D.; Wirth, S.; Bellingrath-Kimura, S.D.; Mishra, J.; Arora, N.K. Salt-tolerant plant growth promoting rhizobacteria for enhancing crop productivity of saline soils. Front. Microbiol. 2019, 10, 2791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Egamberdieva, D.; Shurigin, V.; Alaylar, B.; Wirth, S.; Bellingrath-Kimura, S.D. Bacterial endophytes from horseradish (Armoracia rusticana G. Gaertn.,B.Mey. &Scherb.) with antimicrobial efficacy against pathogens. Plant Soil Environ. 2020, 66, 309–316. [Google Scholar]
- Mukherjee, S.; Das, P.; Sen, R. Towards commercial production of microbial surfactants. Trends. Biotechnol. 2006, 24, 509–515. [Google Scholar] [CrossRef]
- Santos, D.K.; Rufino, R.D.; Luna, J.M.; Santos, V.A.; Sarubbo, L.A. Biosurfactants: Multifunctional biomolecules of the 21st century. Int. J. Mol. Sci. 2016, 17, 401. [Google Scholar] [CrossRef] [Green Version]
- D’aes, J.; De Maeyer, K.; Pauwelyn, E.; Höfte, M. Biosurfactants in plant–Pseudomonas interactions and their importance to biocontrol. Environ. Microbiol. Rep. 2010, 2, 359–372. [Google Scholar] [CrossRef]
- Monnier, N.; Furlan, A.; Botcazon, C.; Dahi, A.; Mongelard, G.; Cordelier, S.; Clément, C.; Dorey, S.; Sarazin, C.; Rippa, S. Rhamnolipids from Pseudomonas aeruginosa are elicitors triggering Brassica napus protection against Botrytis cinerea without physiological disorders. Front. Plant Sci. 2018, 9, 1170. [Google Scholar] [CrossRef] [Green Version]
- Fenibo, E.O.; Ijoma, G.N.; Selvarajan, R.; Chikere, C.B. Microbial surfactants: The next generation multifunctional biomolecules for applications in the petroleum industry and its associated environmental remediation. Microorganisms 2019, 7, 581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sachdev, D.P.; Cameotra, S.S. Biosurfactants in agriculture. Appl. Microbiol. Biotechnol. 2013, 97, 1005–1016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raaijmakers, J.M.; de Bruijn, I.; Nybroe, O.; Ongena, M. Natural functions of lipopeptides from Bacillus and Pseudomonas: More than surfactants and antibiotics. FEMS. Microbiol. Rev. 2010, 34, 1037–1062. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beneduzi, A.; Ambrosini, A.; Passaglia, L.M. Plant growth-promoting rhizobacteria (PGPR): Their potential as antagonists and biocontrol agents. Genet. Mol. Biol. 2012, 35, 1044–1051. [Google Scholar] [CrossRef] [Green Version]
- Cho, S.T.; Chang, H.H.; Egamberdieva, D.; Kamilova, F.; Lugtenberg, B.; Kuo, C.H. Genome analysis of Pseudomonas fluorescens PCL1751: A rhizobacterium that controls root diseases and alleviates salt stress for its plant host. PLoS ONE 2015, 10, e0140231. [Google Scholar] [CrossRef]
- Pacwa-Plociniczak, P.G.A.; Piotrowska-Seget, Z.; Cameotra, S.S. Environmental applications of biosurfactants: Recent advances. Int. J. Mol. Sci. 2015, 12, 633–654. [Google Scholar] [CrossRef]
- Câmara, J.M.D.A.; Sousa, M.A.S.B.; Barros Neto, E.L.; Oliviera, M.C.A. Application of rhamnolipid biosurfactant produced by Pseudomonas aeruginosa in microbial-enhanced oil recovery (MEOR). J. Petrol. Explor. Prod. Technol. 2019, 9, 2333–2341. [Google Scholar] [CrossRef] [Green Version]
- Bhardwaj, A.R.; Joshi, G.; Kukreja, B.; Malik, V.; Arora, P.; Pandey, R.; Shukla, R.N.; Bankar, K.G.; Katiyar-Agarwal, S.; Goel, S.; et al. Global insights into high temperature and drought stress regulated genes by RNA-Seq in economically important oilseed crop Brassica juncea. BMC Plant. Biol. 2015, 2, 15. [Google Scholar] [CrossRef] [Green Version]
- Cartea, M.E.; Francisco, M.; Soengas, P.; Velasco, P. Phenolic compounds in Brassica vegetables. Molecules 2010, 16, 251–280. [Google Scholar] [CrossRef]
- Kapoor, D.; Kaur, S.; Bhardwaj, R. Physiological and biochemical changes in Brassica juncea plants under Cd-Induced stress. Biomed. Res. Int. 2014, 2014, 726070. [Google Scholar] [CrossRef] [Green Version]
- Thakur, A.K.; Chatterjee, S.S.; Kumar, V. Antidepressant-like effects of Brassica juncea L. leaves in diabetic rodents. Indian J. Exp. Biol. 2014, 52, 613–622. [Google Scholar] [PubMed]
- Baux, A.; Hebeisen, T.; Pellet, D. Effects of minimal temperature on low linolenic rapeseed on fatty acid composition. Eur. J. Agron. 2008, 29, 102–107. [Google Scholar] [CrossRef]
- Zhang, Y. Allyl isothiocyanate as a cancer chemopreventive phytochemical. Mol. Nutr. Food Res. 2010, 54, 127–135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, H.K.; Ingle, S.; Singh, C.; Sarkar, B.C.; Upadhyay, A. Effect of various process treatment conditions on the allyl isothiocyanate extraction rate from mustard meal. J. Food. Sci. Technol. 2012, 49, 368–372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iqbal, N. Phytosterols of Brassica juncea (Indian Mustard). Int. J. Res. Rev. 2019, 6, 24–27. [Google Scholar]
- Shekhawat, K.; Rathore, S.S.; Premi, O.P.; Kandpal, B.K.; Chauhan, J.S. Advances in agronomic management of Indian mustard (Brassica juncea (L.) Czernj. Cosson): An overview. Int. J. Agron. 2012, 2012, 408284. [Google Scholar] [CrossRef] [Green Version]
- Jat, R.S.; Singh, V.V.; Sharma, P.; Rai, P.K. Oilseed brassica in India: Demand, supply, policy perspective and future potential. Oilseeds Fats Crops Lipids 2019, 26, 8. [Google Scholar] [CrossRef] [Green Version]
- Garrity, G. The proteobacteria, Part B the gammaproteobacteria. In Bergey’s Manual of Systematic Bacteriology; Garrity, G., Brenner, D.J., Krieg, N.R., Staley, J.R., Eds.; Springer: New York, NY, USA, 2005; Volume 3, pp. 323–337. [Google Scholar]
- Molina, L.; Segura, A.; Duque, E.; Ramos, J.L. The versatility of Pseudomonas putida in the rhizosphere environment. Adv. Appl. Microbiol. 2020, 110, 149–180. [Google Scholar]
- Alsaim, H.A.A. Isolation and characterization of phosphate solubilizing Pseudomonas species and assess its efficacy as plant growth promoter. Biochem. Cell. Arch. 2020, 20, 2301–2308. [Google Scholar]
- Oteino, N.; Lally, R.D.; Kiwanuka, S.; Lloyd, A.; Ryan, D.; Germaine, K.J.; Dowling, D.N. Plant growth promotion induced by phosphate solubilizing endophytic Pseudomonas isolates. Front. Microbiol. 2015, 6, 745. [Google Scholar] [CrossRef] [Green Version]
- Baliah, N.T.; Pandiarajan, G.; Kumar, B.M. Isolation, identification and characterization of phosphate solubilizing bacteria from different crop soils of Srivilliputtur Taluk, Virudhunagar District, Tamil Nadu. Trop. Ecol. 2016, 57, 465–474. [Google Scholar]
- Alori, E.T.; Glick, B.R.; Babalola, O.O. Microbial phosphorus solubilization and its potential for use in sustainable agriculture. Front. Microbiol. 2017, 8, 971. [Google Scholar] [CrossRef] [Green Version]
- Sarikhani, M.R.; Aliasgharzad, N.; Khoshru, B. P solubilizing potential of some plant growth promoting bacteria used as ingredient in phosphatic biofertilizers with emphasis on growth promotion of Zea mays L. Geomicrobiol. J. 2020, 37, 327–335. [Google Scholar] [CrossRef]
- Srivastava, S.; Srivastava, S. Prescience of endogenous regulation in Arabidopsis thaliana by Pseudomonas putida MTCC 5279 under phosphate starved salinity stress condition. Sci. Rep. 2020, 10, 5855. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patten, C.L.; Glick, B.R. Role of Pseudomonas putida indoleacetic acid in development of the host plant root system. Appl. Environ. Microbiol. 2002, 68, 3795–3801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- da Silva, M.J.C.; Junior, S.F.P.; Junior, K.F.; Nascimento, V.X.; de Medeiros, A.S.; da Silva, S.J.C.; Alves, M.M.S.; Sant’Ana, A.E.G. IAA production of indigenous isolate of plant growth promoting rhizobacteria in the presence of tryptophan. Aust. J. Crop. Sci. 2020, 14, 537–544. [Google Scholar] [CrossRef]
- Habibi, S.; Djedidi, S.; Ohkama-Ohtsu, N.; Sarhadi, W.A.; Kojima, K.; Rallos, R.V.; Ramirez, M.; Yamaya, H.; Sekimoto, H.; Yokoyama, T. Isolation and screening of indigenous plant growth-promoting rhizobacteria from different rice cultivars in Afghanistan soils. Microbes Environ. 2019, 34, 347–355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hernández-Montiel, L.G.; Contreras, C.J.C.; Amador, B.M.; Hernández, L.V.; Aguilar, E.E.Q.; Contreras, R.G.C. Efficiency of two inoculation methods of Pseudomonas putida on growth and yield of tomato plants. J. Soil Sci. Plant Nutr. 2017, 17, 1003–1012. [Google Scholar] [CrossRef] [Green Version]
- Hussein, K.A.; Joo, J.H. Stimulation, purification, and chemical characterization of siderophores produced by the rhizospheric bacterial strain Pseudomonas putida. Rhizosphere 2017, 4, 16–21. [Google Scholar] [CrossRef]
- Sayyed, R.Z.; Badgujar, M.D.; Sonawane, H.M.; Mhaske, M.M.; Chincholkar, S.B. Production of microbial iron chelators (siderophores) by fluorescent pseudomonads. Indian J. Biotechnol. 2005, 4, 484–490. [Google Scholar]
- Ferreira, M.J.; Silva, H.; Cunha, A. Siderophore-producing Rhizobacteria as a promising tool for empowering plants to cope with iron limitation in saline soils: A review. Pedosphere 2019, 29, 409–420. [Google Scholar] [CrossRef]
- Barrientos-Moreno, L.; Molina-Henares, M.A.; Pastor-García, M.; Ramos-González, M.I.; Espinosa-Urgel, M. Arginine biosynthesis modulates pyoverdine production and release in Pseudomonas putida as part of the mechanism of adaptation to oxidative stress. J. Bacteriol. 2019, 201, e00454-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, P.; Trivedi, G.; Saraf, M. Iron biofortification in mungbean using siderophore producing plant growth promoting bacteria. Environ. Sustain. 2018, 1, 357–365. [Google Scholar] [CrossRef]
- Sharma, R.; Singh, J.; Verma, N. Optimization of rhamnolipid production from Pseudomonas aeruginosa PBS towards application for microbial enhanced oil recovery. 3 Biotech 2018, 8, 20. [Google Scholar] [CrossRef]
- Joe, M.M.; Gomathi, R.; Benson, A.; Shalini, D.; Rengamsamy, P.; Henry, A.; Truu, J.; Truu, M.; Sa, T. Simultaneous application of biosurfactant and bioaugmentation with rhamnolipid-producing shewanella for enhanced bioremediation of oil-polluted soil. Appl. Sci. 2019, 9, 3773. [Google Scholar] [CrossRef] [Green Version]
- Eraqi, W.A.; Yassin, A.S.; Ali, A.E.; Amin, M.A. Utilization of crude glycerol as a substrate for the production of rhamnolipid by Pseudomonas aeruginosa. Biotechnol. Res. Int. 2016, 2016, 3464509. [Google Scholar] [CrossRef] [Green Version]
- Hassen, W.; Neifar, M.; Cherif, H.; Najjari, A.; Chouchane, H.; Driouich, R.C.; Salah, A.; Naili, F.; Mosbah, A.; Souissi, Y.; et al. Pseudomonas rhizophila S211, a new plant growth-promoting rhizobacterium with potential in pesticide-bioremediation. Front. Microbiol. 2018, 9, 34. [Google Scholar] [CrossRef] [Green Version]
- Olasanmi, I.O.; Thring, R.W. The role of biosurfactants in the continued drive for environmental sustainability. Sustainability 2018, 10, 4817. [Google Scholar] [CrossRef] [Green Version]
- Jarvis, F.G.; Johnson, M.J. A glycolipid produced by Pseudomonas aeruginosa. J. Am. Chem. Soc. 1949, 71, 4124–4126. [Google Scholar] [CrossRef]
- Gunther, N.W.; Nunez, A.; Fett, W.; Solaiman, D.K.Y. Production of rhamnolipids by Pseudomonas chlororaphis, a nonpathogenic bacterium. Appl. Environ. Microbiol. 2005, 71, 2288–2293. [Google Scholar] [CrossRef] [Green Version]
- Abdel-Mawgoud, A.M.; Lépinem, F.; Déziel, E. Rhamnolipids: Diversity of structures, microbial origins and roles. Appl. Microbiol. Biotechnol. 2010, 86, 1323–1336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sekhon Randhawa, K.K.; Rahman, P.K. Rhamnolipid biosurfactants-past, present, and future scenario of global market. Front. Microbiol. 2014, 5, 454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kruijt, M.; Tran, H.; Raaijmakers, J.M. Functional, genetic and chemical characterization of biosurfactants produced by plant growth-promoting Pseudomonas putida 267. J. Appl. Microbiol. 2009, 107, 546–556. [Google Scholar] [CrossRef]
- Ángeles, M.T.; Refugio, R.V. In situ biosurfactant production and hydrocarbon removal by Pseudomonas putida CB-100 in bioaugmented and biostimulated oil-contaminated soil. Braz. J. Microbiol. 2013, 44, 595–605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernat, P.; Nesme, J.; Paraszkiewicz, K.; Schloter, M.; Płaza, G. Characterization of extracellular biosurfactants expressed by a Pseudomonas putida strain isolated from the interior of healthy roots from Sida hermaphrodita grown in a heavy metal contaminated soil. Curr. Microbiol. 2019, 76, 1320–1329. [Google Scholar] [CrossRef] [PubMed]
- Heyd, M.; Kohnert, A.; Tan, T.H.; Nusser, M.; Kirschhöfer, F.; Brenner-Weiss, G.; Franzreb, M.; Berensmeier, S. Development and trends of biosurfactant analysis and purification using rhamnolipids as an example. Anal. Bioanal. Chem. 2008, 391, 1579–1590. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Tiwary, B.N. Isolation and characterization of glycolipid biosurfactant produced by a Pseudomonas otitidis strain isolated from Chirimiri coal mines, India. Bioresour. Bioprocess. 2016, 3, 42. [Google Scholar] [CrossRef] [Green Version]
- Kiefer, J.; Radzuan, M.N.; Winterburn, J. Infrared spectroscopy for studying structure and aging effects in rhamnolipid biosurfactants. Appl. Sci. 2017, 7, 533. [Google Scholar] [CrossRef]
- Leitermann, F.; Syldatk, C.; Hausmann, R. Fast quantitative determination of microbial rhamnolipids from cultivation broths by ATR-FTIR Spectroscopy. J. Biol. Eng. 2008, 2, 13. [Google Scholar] [CrossRef] [Green Version]
- Günzler, H.; Gremlich, H.-U. IR-Spektroskopie; Wiley-VCH: Weinhein, Germany, 2003; Volume 1, p. 365. [Google Scholar]
- Mnif, I.; Ellouz-Chaabouni, S.; Ghribi, D. Glycolipid biosurfactants, main classes, functional properties and related potential applications in environmental biotechnology. J. Polym. Environ. 2018, 26, 2192–2206. [Google Scholar] [CrossRef]
- Mnif, I.; Ghribi, D. Glycolipid biosurfactants: Main properties and potential applications in agriculture and food industry. J. Sci. Food Agric. 2016, 96, 4310–4320. [Google Scholar] [CrossRef] [PubMed]
- Akbari, S.; Abdurahman, N.H.; Yunus, R.M.; Fayaz, F.; Alara, O.R. Biosurfactants; a new frontier for social and environmental safety: A mini review. Biotechnol. Res. Innov. 2018, 2, 81–90. [Google Scholar] [CrossRef]
- Zhang, L.; Veres-Schalnat, T.A.; Somogyi, A.; Pemberton, J.E.; Maier, R.M. Fatty acid cosubstrates provide β-oxidation precursors for rhamnolipid biosynthesis in Pseudomonas aeruginosa, as evidenced by isotope tracing and gene expression assays. Appl. Environ. Microbiol. 2012, 78, 8611–8622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bajpai, A.; Singh, B.; Johri, B.N. Rhamnolipids and siderophores from Pseudomonas protegens strain BNJ-SS-45 isolated from wheat rhizosphere. Environ. Sustain. 2020, 3, 219–228. [Google Scholar] [CrossRef]
- Buonocore, C.; Tedesco, P.; Vitale, G.A.; Palma Esposito, F.; Giugliano, R.; Monti, M.C.; D’Auria, M.V.; de Pascale, D. Characterization of a new mixture of mono-rhamnolipids produced by Pseudomonas gessardii isolated from Edmonson Point (Antarctica). Mar. Drugs 2020, 18, 269. [Google Scholar] [CrossRef]
- Sabturani, N.; Latif, J.; Radiman, S.; Hamzah, A. Spectroscopic analysis of rhamnolipid produced by produced by Pseudomonas aeruginosa UKMP14T. Malays. J. Anal. Sci. 2016, 20, 31–43. [Google Scholar] [CrossRef]
- Loiseau, C.; Portier, E.; Corre, M.; Schlusselhuber, M.; Depayras, S.; Berjeaud, J.; Verdon, J. Highlighting the potency of biosurfactants produced by Pseudomonas strains as anti-Legionella agents. Biomed. Res. Int. 2018, 2018, 8194368. [Google Scholar] [CrossRef] [Green Version]
- Tremblay, J.; Richardson, A.P.; Lépine, F.; Déziel, E. Self-produced extracellular stimuli modulate the Pseudomonas aeruginosa swarming motility behaviour. Environ. Microbiol. 2007, 9, 2622–2630. [Google Scholar] [CrossRef]
- Sinumvayo, J.P.; Ishimwe, N. Agriculture and food applications of rhamnolipids and its production by Pseudomonas aeruginosa. J. Chem. Eng. Process. Technol. 2015, 6, 223. [Google Scholar] [CrossRef] [Green Version]
- Dusane, D.; Rahman, P.; Zinjarde, S.; Venugopalan, V.; McLean, R.; Weber, M. Quorum sensing; implication on rhamnolipid biosurfactant production. Biotechnol. Genet. Eng. Rev. 2010, 27, 159–184. [Google Scholar] [CrossRef] [Green Version]
- Singh, R.; Glick, B.R.; Rathore, D. Biosurfactants as a biological tool to increase micronutrient availability in soil: A review. Pedosphere 2018, 28, 170–189. [Google Scholar] [CrossRef]
- Plaza, G.; Achal, V. Biosurfactants: Eco-Friendly and Innovative Biocides against Biocorrosion. Int. J. Mol. Sci. 2020, 21, 2152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Awada, S.M.; Awada, M.M.; Spendlove, R.S. Method of Controlling Pests with Biosurfactant Penetrants as Carriers for Active Agents. U.S. Patent US20110319341A1, 2011. Available online: https://patents.google.com/patent/US20110319341A1/en (accessed on 9 August 2020).
- Thavasi, R.; Marchant, R.; Banat, I.M. Biosurfactant applications in agriculture. In Bisurfactants: Production and Utilization-Processes, Technologies and Economics; Kosaric, N., Vardar, F., Eds.; CRC Press: Boca Raton, VA, USA, 2014; Volume 159, p. 389. [Google Scholar]
- Chaprão, M.J.; Ferreira, I.N.S.; Correa, P.F.; Rufino, R.D.; Luna, J.M.; Silva, E.J.; Sarubbo, L.A. Application of bacterial and yeast biosurfactants for enhanced removal and biodegradation of motor oil from contaminated sand. Electron. J. Biotechnol. 2015, 18, 471–479. [Google Scholar] [CrossRef] [Green Version]
- Srinivasan, R.; Karaoz, U.; Volegova, M.; MacKichan, J.; Kato-Maeda, M.; Miller, S.; Nadarajan, R.; Brodie, E.L.; Lynch, S.V. Use of 16S rRNA gene for identification of a broad range of clinically relevant bacterial pathogens. PLoS ONE 2015, 10, e0117617. [Google Scholar] [CrossRef] [PubMed]
- Pikovskaya, R.I. Mobilization of phosphorus in soil connection with the vital activity of some microbial species. Microbiology 1948, 17, 362–370. [Google Scholar]
- Ahmad, F.; Ahmad, I.; Khan, M.S. Screening of free-living rhizospheric bacteria for their multiple plant growth promoting activities. Microbiol. Res. 2008, 163, 173–181. [Google Scholar] [CrossRef] [PubMed]
- Schwyn, B.; Neilands, J.B. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef]
- Arora, N.K.; Verma, M. Modified microplate method for rapid and efficient estimation of siderophore produced by bacteria. 3 Biotech 2017, 7, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Mulligan, C.N.; Cooper, D.G.; Neufeld, R.J. Selection of microbes producing biosurfactants in media without hydrocarbons. J. Ferment. Technol. 1984, 62, 311–314. [Google Scholar]
- Morikawa, M.; Hirata, Y.; Imanaka, T. A study on the structure-function relationship of the lipopeptide biosurfactant. Biochim. Biophys. Acta 2000, 1488, 211–218. [Google Scholar] [CrossRef]
- Siegmund, I.; Wagner, F. New method for detecting rhamnolipids excreted by Pseudomonas species during growth on mineral agar. Biotechnol. Tech. 1991, 5, 265–268. [Google Scholar] [CrossRef]
- Cooper, D.; Goldenberg, B. Surface-active agents from 2 Bacillus species. Appl. Environ. Microbiol. 1987, 53, 224–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandes, P.A.V.; Arruda, I.R.; Santos, A.F.A.B.; Araujo, A.A.; Maior, A.A.S.; Ximenes, E.A. Antimicrobial activity of surfactants produced by Bacillus subtilis R14 against multidrug-resistant bacteria. Braz. J. Microbiol. 2007, 38, 704–709. [Google Scholar] [CrossRef] [Green Version]
- Shreve, G.S.; Makula, R. Characterization of a new rhamnolipid biosurfactant complex from Pseudomonas isolate dyna270. Biomolecules 2019, 9, 885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali, N.; Wang, F.; Xu, B.; Safdar, B.; Ullah, A.; Naveed, M.; Wang, C.; Rashid, M.T. Production and application of biosurfactant produced by Bacillus licheniformis Ali5 in enhanced oil recovery and motor oil removal from contaminated sand. Molecules 2019, 24, 4448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haba, E.; Abalos, A.; Jáuregui, O.; Espuny, M.J.; Manresa, A. Use of liquid chromatography–mass spectroscopyfor studying the composition and propertiesof rhamnolipids produced by different strains of Pseudomonas aeruginosa. J. Surfactants Deterg. 2003, 20, 155–161. [Google Scholar] [CrossRef]
- Jackson, M.L. Soil Chemical Analysis; Prentice Hall of India Pvt.Ltd.: New Delhi, India, 1973; pp. 38–56. [Google Scholar]
- Ge, S.; Zhu, Z.; Peng, L.; Chen, Q.; Jiang, Y. Soil nutrient status and leaf nutrient diagnosis in the main apple producing regions in China. Hortic. Plant. J. 2018, 4, 89–93. [Google Scholar] [CrossRef]
- Bhanu, A.N.; Srivastava, K.; Singh, R.K. Advances in agronomic management in Indian mustard for Eastern Uttar Pradesh. Acta Sci. Agric. 2019, 3, 70–79. [Google Scholar] [CrossRef]
- Nandakumar, R.; Babu, S.; Viswanathan, R.; Sheela, J.; Raguchander, T.; Samiyappan, R. A new bio-formulation containing plant growth promoting rhizobacterial mixture for the management of sheath blight and enhanced grain yield in rice. BioControl 2001, 46, 493–510. [Google Scholar] [CrossRef]
- Fatima, T.; Mishra, I.; Verma, R.; Arora, N.K. Mechanisms of halotolerant plant growth promoting Alcaligenes sp. involved in salt tolerance and enhancement of the growth of rice under salinity stress. 3Biotech 2020, 10, 361. [Google Scholar] [CrossRef]
- Arnon, D.I. Copper enzymes in isolated chloroplasts, polyphenoxidase in Beta vulgaris. Plant. Physiol. 1949, 24, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, C.C.; Yang, M.H.; Wen, H.M.; Chern, J.C. Estimation of total flavonoid content in propolis by two complementary colorimetric methods. J. Food Drug Anal. 2002, 10, 178–182. [Google Scholar]




| Treatment | % Germination | Root Length (cm) | Shoot Length (cm) | Total Fresh Weight (gm) | Total Dry Weight (gm) | Number of Pods | Total Oil Content (%) * | Total Chlorophyll Content (mg/g) | Total Flavonoid Content (mg/g) |
|---|---|---|---|---|---|---|---|---|---|
| C | 50.1 ± 0.5 a | 15 ± 0.4 a | 119.3 ± 0.5 a | 290.8 ± 1.4 a | 114 ± 1.02 a | 284.2 ± 8.7 a | 35.0% | 16.6 ± 0.1 a | 6.4 ± 0.2 a |
| T1 | 62.3 ± 0.9 b | 21.9 ± 0.1 b | 179.7 ± 1.3 b | 323.4 ± 4.2 b | 129.2 ± 1.9 b | 379.8 ± 11.3 b | 39.0% | 23.4 ± 0.16 b | 7.5 ± 0.2 b |
| T2 | 71.5 ± 0.6 c | 24.6 ± 0.3 c | 191.3 ± 1.4 c | 354.5 ± 2.9 c | 149.2 ± 0.8 c | 394 ± 4.7 b | 40.5% | 25.2 ± 0.1 c | 8.5 ± 0.1 c |
| T3 | 84.9 ± 0.6 d | 26.1 ± 0.5 de | 198.8 ± 0.9 e | 374.2 ± 2.1 d | 164.5 ± 1.1 d | 426.8 ± 1.2 c | 41.5% | 28.5 ± 0.2 e | 10.5 ± 0.2 e |
| T4 | 83.9 ± 0.9 d | 25.2 ± 0.4 cd | 195.4 ± 0.5 d | 371.2 ± 1.9 d | 161 ± 1.4 d | 421.6 ± 2.8 c | 41.0% | 27.2 ± 0.2 d | 9.5 ± 0.2 d |
| T5 | 88.2 ± 0.7 e | 28.1 ± 0.1 f | 213.8 ± 1.1 g | 405.5 ± 1.7 e | 197.0 ± 1.2 e | 429.4 ± 6.6 c | 43.5% | 31.4 ± 0.2 f | 12.2 ± 0.2 g |
| T6 | 91.3 ± 0.2 f | 30.1 ± 0.1 g | 216 ± 1.8 g | 483.4 ± 5.3 f | 204 ± 4.6 e | 481.2 ± 2.1 d | 45.0% | 32.3 ± 0.1 h | 13.5 ± 0.1 h |
| T7 | 85.2 ± 1.1 d | 27.4 ± 0.5 ef | 202.8 ± 0.7 f | 379 ± 3.2 d | 168.6 ± 1.3 d | 427 ± 4.2 c | 42.5% | 30.4 ± 0.1 g | 11.2 ± 0.1 f |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mishra, I.; Fatima, T.; Egamberdieva, D.; Arora, N.K. Novel Bioformulations Developed from Pseudomonas putida BSP9 and Its Biosurfactant for Growth Promotion of Brassica juncea (L.). Plants 2020, 9, 1349. https://0-doi-org.brum.beds.ac.uk/10.3390/plants9101349
Mishra I, Fatima T, Egamberdieva D, Arora NK. Novel Bioformulations Developed from Pseudomonas putida BSP9 and Its Biosurfactant for Growth Promotion of Brassica juncea (L.). Plants. 2020; 9(10):1349. https://0-doi-org.brum.beds.ac.uk/10.3390/plants9101349
Chicago/Turabian StyleMishra, Isha, Tahmish Fatima, Dilfuza Egamberdieva, and Naveen Kumar Arora. 2020. "Novel Bioformulations Developed from Pseudomonas putida BSP9 and Its Biosurfactant for Growth Promotion of Brassica juncea (L.)" Plants 9, no. 10: 1349. https://0-doi-org.brum.beds.ac.uk/10.3390/plants9101349