The Versatile Roles of Sulfur-Containing Biomolecules in Plant Defense—A Road to Disease Resistance
Abstract
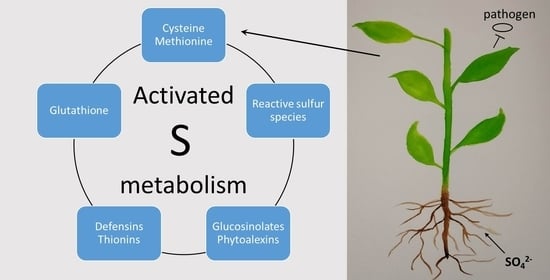
:1. Introduction
2. Sulfur Containing Amino Acids (SAAs) in Plant Disease Resistance
2.1. Cysteine
2.2. Methionine
3. Glutathione (GSH) in Plant Disease Resistance
3.1. GSH Correlates with Plant Resistance
3.2. Artificial Modification of GSH Levels in Plants Affects Disease Resistance
3.3. GSH and Plant Hormones
3.3.1. GSH and SA
3.3.2. GSH and Jasmonic Acid
3.3.3. GSH and Ethylene
3.4. Glutathione S-Transferases
4. Sulfur Containing Pathogenesis Related (PR) Antimicrobial Peptides (AMPs) in Plant Disease Resistance
5. Sulfur-Containing Secondary Metabolites (Phytoalexins, Phytoanticipins) in Plant Disease Resistance
5.1. Sulfur-Containing Phytoalexins
5.2. Phytoanticipins
5.2.1. Glucosinolates
5.2.2. Thiosulfinates
6. Reactive Sulfur Species (RSS)
7. Conclusions and Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Schnug, E.; Haneklaus, S. Diagnosis of sulphur nutrition. In Sulphur in Agroecosystems. Nutrients in Ecosystems; Schnug, E., Ed.; Springer: Dordrecht, The Netherlands, 1998; Volume 2, pp. 1–38. ISBN 978-94-011-5100-9. [Google Scholar]
- Ryant, P.; Dolezelova, E.; Fabrik, I.; Baloun, J.; Adam, V.; Babula, P.; Kizek, R. Electrochemical determination of low molecular mass thiols content in potatoes (Solanum tuberosum) cultivated in the presence of various sulphur forms and infected by late blight (Phytophora infestans). Sensors 2008, 8, 3165–3182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bloem, E.; Riemenschneider, A.; Volker, J.; Papenbrock, J.; Schmidt, A.; Salac, I.; Haneklaus, S.; Schnug, E. Sulphur supply and infection with Pyrenopeziza brassicae influence L-cysteine desulphydrase activity in Brassica napus L. J. Exp. Bot. 2004, 55, 2305–2312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bloem, E.; Haneklaus, S.; Salac, I.; Wickenhäuser, P.; Schnug, E. Facts and fiction about sulfur metabolism in relation to plant-pathogen interactions. Plant Biol. 2007, 9, 596–607. [Google Scholar] [CrossRef] [PubMed]
- Kruse, C.; Jost, R.; Lipschis, M.; Kopp, B.; Hartmann, M.; Hell, R. Sulfur-enhanced defence: Effects of sulfur metabolism, nitrogen supply, and pathogen lifestyle. Plant Biol. 2007, 9, 608–619. [Google Scholar] [CrossRef]
- Dubuis, P.H.; Marazzi, C.; Städler, E.; Mauch, F. Sulphur deficiency causes a reduction in antimicrobial potential and leads to increased disease susceptibility of oilseed rape. J. Phytopathol. 2005, 153, 27–36. [Google Scholar] [CrossRef] [Green Version]
- Bloem, E.; Haneklaus, S.; Schnug, E. Milestones in plant sulfur research on sulfur-induced-resistance (SIR) in Europe. Front. Plant Sci. 2015, 5, 779. [Google Scholar] [CrossRef]
- Kruse, C.; Haas, F.H.; Jost, R.; Reiser, B.; Reichelt, M.; Wirtz, M.; Gershenzon, J.; Schnug, E.; Hell, R. Improved sulfur nutrition provides the basis for enhanced production of sulfur-containing defense compounds in Arabidopsis thaliana upon inoculation with Alternaria brassicicola. J. Plant Physiol. 2012, 169, 740–743. [Google Scholar] [CrossRef]
- Capaldi, F.R.; Gratão, P.L.; Reis, A.R.; Lima, L.W.; Azevedo, R.A. Sulfur metabolism and stress defense responses in plants. Trop. Plant Biol. 2015, 8, 60–73. [Google Scholar] [CrossRef] [Green Version]
- Aarabi, F.; Naake, T.; Fernie, A.R.; Hoefgen, R. Coordinating sulfur pools under sulfate deprivation. Trends Plant Sci. 2020, 25, 1227–1239. [Google Scholar] [CrossRef]
- Hawkesford, M.; Horst, W.; Kichey, T.; Lambers, H.; Schjoerring, J.; Møller, I.S.; White, P. Functions of macronutrients. In Marschner’s Mineral Nutrition of Higher Plants, 3rd ed.; Marschner, P., Ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2011; pp. 135–189. ISBN 9780123849052. [Google Scholar]
- Takahashi, H.; Kopriva, S.; Giordano, M.; Saito, K.; Hell, R. Sulfur assimilation in photosynthetic organisms: Molecular functions and regulations of transporters and assimilatory enzymes. Annu. Rev. Plant Biol. 2011, 62, 157–184. [Google Scholar] [CrossRef]
- Iqbal, N.; Masood, A.; Khan, M.I.R.; Asgher, M.; Fatma, M.; Khan, N.A. Cross-talk between sulfur assimilation and ethylene signaling in plants. Plant Signal. Behav. 2013, 8, e22478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gigolashvili, T.; Kopriva, S. Transporters in plant sulfur metabolism. Front. Plant Sci. 2014, 5, 442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prioretti, L.; Gontero, B.; Hell, R.; Giordano, M. Diversity and regulation of ATP sulfurylase in photosynthetic organisms. Front. Plant Sci. 2014, 5, 597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leustek, T.; Saito, K. Sulfate transport and assimilation in plants. Plant Physiol. 1999, 120, 637–643. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, M.; Hubberten, H.M.; Saito, K.; Hoefgen, R. General regulatory patterns of plant mineral nutrient depletion as revealed by serat quadruple mutants disturbed in cysteine synthesis. Mol. Plant 2010, 3, 438–466. [Google Scholar] [CrossRef]
- Leustek, T.; Martin, M.N.; Bick, J.A.; Davies, J.P. Pathways and regulation of sulfur metabolism revealed through molecular and genetic studies. Annu. Rev. Plant Biol. 2000, 51, 141–165. [Google Scholar] [CrossRef]
- Saito, K. Regulation of sulfate transport and synthesis of sulfur-containing amino acids. Curr. Opin. Plant Biol. 2000, 3, 188–195. [Google Scholar] [CrossRef]
- Nakai, Y.; Maruyama-Nakashita, A. Biosynthesis of sulfur-containing small biomolecules in plants. Int. J. Mol. Sci. 2020, 21, 3470. [Google Scholar] [CrossRef]
- Álvarez, C.; Bermúdez, M.Á.; Romero, L.C.; Gotor, C.; García, I. Cysteine homeostasis plays an essential role in plant immunity. New Phytol. 2012, 193, 165–177. [Google Scholar] [CrossRef] [Green Version]
- Davidian, J.-C.; Kopriva, S. Regulation of sulfate uptake and assimilation—The same or not the same? Mol. Plant 2010, 3, 314–325. [Google Scholar] [CrossRef]
- Dong, Y.; Silbermann, M.; Speiser, A.; Forieri, I.; Linster, E.; Poschet, G.; Allboje Samami, A.; Wanatabe, M.; Sticht, C.; Teleman, A.A.; et al. Sulfur availability regulates plant growth via glucose-TOR signaling. Nat. Commun. 2017, 8, 1174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abadie, C.; Tcherkez, G. Plant sulphur metabolism is stimulated by photorespiration. Commun. Biol. 2019, 2, 379. [Google Scholar] [CrossRef] [PubMed]
- Maruyama-Nakashita, A.; Nakamura, Y.; Tohge, T.; Saito, K.; Takahashi, H. Arabidopsis SLIM1 is a central transcriptional regulator of plant sulfur response and metabolism. Plant Cell 2006, 18, 3235–3251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yi, H.; Galant, A.; Ravilious, G.E.; Preuss, M.L.; Jez, J.M. Sensing sulfur conditions: Simple to complex protein regulatory mechanisms in plant thiol metabolism. Mol. Plant 2010, 3, 269–279. [Google Scholar] [CrossRef]
- Joshi, N.C.; Meyer, A.J.; Bangash, S.A.K.; Zheng, Z.; Leustek, T. Arabidopsis γ-glutamylcyclotransferase affects glutathione content and root system architecture during sulfur starvation. New Phytol. 2019, 221, 1387–1397. [Google Scholar] [CrossRef] [Green Version]
- Aarabi, F.; Kusajima, M.; Tohge, T.; Konishi, T.; Gigolashvili, T.; Takamune, M.; Sasazaki, Y.; Watanabe, M.; Nakashita, H.; Fernie, A.R.; et al. Sulfur deficiency-induced repressor proteins optimize glucosinolate biosynthesis in plants. Sci. Adv. 2016, 2, e1601087. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.-Y.; Chao, D.-Y.; Koprivova, A.; Danku, J.; Wirtz, M.; Müller, S.; Sandoval, F.J.; Bauwe, H.; Roje, S.; Dilkes, B.; et al. Nuclear localised more sulphur ACCUMULATION1 epigenetically regulates sulphur homeostasis in Arabidopsis thaliana. PLoS Genet. 2016, 12, e1006298. [Google Scholar] [CrossRef]
- Rausch, T.; Wachter, A. Sulfur metabolism: A versatile platform for launching defence operations. Trends Plant Sci. 2005, 10, 503–509. [Google Scholar] [CrossRef]
- Romero, L.C.; Aroca, M.Á.; Laureano-Marín, A.M.; Moreno, I.; García, I.; Gotor, C. Cysteine and cysteine-related signaling pathways in Arabidopsis thaliana. Mol. Plant 2014, 7, 264–276. [Google Scholar] [CrossRef] [Green Version]
- Gotor, C.; Laureano-Marín, A.M.; Moreno, I.; Aroca, Á.; García, I.; Romero, L.C. Signaling in the plant cytosol: Cysteine or sulfide? Amino Acids 2015, 47, 2155–2164. [Google Scholar] [CrossRef] [Green Version]
- Couturier, J.; Chibani, K.; Jacquot, J.P.; Rouhier, N. Cysteine-based redox regulation and signaling in plants. Front. Plant Sci. 2013, 4, 105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vogelsang, L.; Dietz, K.J. Regulatory thiol oxidation in chloroplast metabolism, oxidative stress response and environmental signaling in plants. Biochem. J. 2020, 447, 1865–1878. [Google Scholar] [CrossRef] [PubMed]
- Jost, R.; Altschmied, L.; Bloem, E.; Bogs, J.; Gershenzon, J.; Hähnel, U.; Hänsch, R.; Hartmann, T.; Kopriva, S.; Kruse, C.; et al. Expression profiling of metabolic genes in response to methyl jasmonate reveals regulation of genes of primary and secondary sulfur-related pathways in Arabidopsis thaliana. Photosynth. Res. 2005, 86, 491–508. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.H.; Hu, K.D.; Hu, L.Y.; Li, Y.H.; Hu, L.B.; Yan, H.; Liu, Y.S.; Zhang, H. An antifungal role of hydrogen sulfide on the postharvest pathogens Aspergillus niger and Penicillium italicum. PLoS ONE 2014, 9, e104206. [Google Scholar] [CrossRef] [Green Version]
- Fotopoulos, V.; Christou, A.; Antoniou, C.; Manganaris, G.A. Hydrogen sulphide: A versatile tool for the regulation of growth and defence responses in horticultural crops. J. Hortic. Sci. Biotechnol. 2015, 90, 227–234. [Google Scholar] [CrossRef]
- Tahir, J.; Watanabe, M.; Jing, H.C.; Hunter, D.A.; Tohge, T.; Nunes-Nesi, A.; Brotman, Y.; Fernie, A.R.; Hoefgen, R.; Dijkwel, P.P. Activation of R-mediated innate immunity and disease susceptibility is affected by mutations in a cytosolic O-acetylserine (thiol) lyase in Arabidopsis. Plant J. 2013, 73, 118–130. [Google Scholar] [CrossRef]
- Wilkinson, S.W.; Mageroslashy, M.H.; Sanchez, A.L.; Smith, L.M.; Furci, L.; Cotton, T.E.A.; Krokene, P.; Ton, J. Surviving in a hostile world: Plant strategies to resist pests and diseases. Annu. Rev. Phytopathol. 2019, 57, 505–529. [Google Scholar] [CrossRef] [Green Version]
- Yadeta, K.A.; Elmore, J.M.; Creer, A.Y.; Feng, B.; Franco, J.Y.; Rufian, J.S.; He, P.; Phinney, B.; Coaker, G. A cysteine-rich protein kinase associates with a membrane immune complex and the cysteine residues are required for cell death. Plant Physiol. 2017, 173, 771–787. [Google Scholar] [CrossRef] [Green Version]
- Gu, J.; Sun, J.; Liu, N.; Sun, X.; Liu, C.; Wu, L.; Liu, G.; Zeng, F.; Hou, C.; Han, S.; et al. A novel cysteine-rich receptor-like kinase gene, TaCRK2, contributes to leaf rust resistance in wheat. Mol. Plant Pathol. 2020, 21, 732–746. [Google Scholar] [CrossRef] [Green Version]
- Smirnoff, N.; Arnaud, D. Hydrogen peroxide metabolism and functions in plants. New Phytol. 2019, 221, 1197–1214. [Google Scholar] [CrossRef]
- Wu, F.; Chi, Y.; Jiang, Z.; Xu, Y.; Xie, L.; Huang, F.; Wan, D.; Ni, J.; Yuan, F.; Wu, X.; et al. Hydrogen peroxide sensor HPCA1 is an LRR receptor kinase in Arabidopsis. Nature 2020, 578, 577–581. [Google Scholar] [CrossRef] [PubMed]
- Roblin, G.; Octave, S.; Faucher, M.; Fleurat-Lessard, P.; Berjeaud, J.M. Cysteine: A multifaceted amino acid involved in signaling, plant resistance and antifungal development. Plant Physiol. Biochem. 2018, 129, 77–89. [Google Scholar] [CrossRef] [PubMed]
- Leroux, P. Effect of pH, amino acids and various organic compounds on the fungitoxicity of pyrimethanil, glufosinate, captafol, cymoxanil and fenpiclonil in Botrytis cinerea. Agronomie 1994, 14, 541–554. [Google Scholar] [CrossRef] [Green Version]
- Octave, S.; Amborabé, B.E.; Luini, E.; Ferreira, T.; Fleurat-Lessard, P.; Roblin, G. Antifungal effects of cysteine towards Eutypa lata, a pathogen of vineyards. Plant Physiol. Biochem. 2005, 43, 1006–1013. [Google Scholar] [CrossRef] [PubMed]
- Pivato, M.; Fabrega-Prats, M.; Masi, A. Low-molecular-weight thiols in plants: Functional and analytical implications. Arch. Biochem. Biophys. 2014, 560, 83–99. [Google Scholar] [CrossRef] [PubMed]
- Rahikainen, M.; Alegre, S.; Trotta, A.; Pascual, J.; Kangasjärvi, S. Trans-methylation reactions in plants: Focus on the activated methyl cycle. Physiol. Plant. 2018, 162, 162–176. [Google Scholar] [CrossRef] [PubMed]
- Sarosh, B.R.; Sivaramakrishnan, S.; Shetty, H.S. Elicitation of defense related enzymes and resistance by l-methionine in pearl millet against downy mildew disease caused by Sclerospora graminicola. Plant Physiol. Biochem. 2005, 43, 808–815. [Google Scholar] [CrossRef]
- Boubakri, H.; Wahab, M.A.; Chong, J.; Gertz, C.; Gandoura, S.; Mliki, A.; Bertsch, C.; Soustre-Gacougnolle, I. Methionine elicits H2O2 generation and defense gene expression in grapevine and reduces Plasmopara viticola infection. J. Plant Physiol. 2013, 170, 1561–1568. [Google Scholar] [CrossRef]
- Ludmerszki, E.; Almási, A.; Rácz, I.; Szigeti, Z.; Solti, Á.; Oláh, C.; Rudnóy, S. S-methylmethionine contributes to enhanced defense against Maize dwarf mosaic virus infection in maize. Rev. Bras. Bot. 2015, 38, 771–782. [Google Scholar] [CrossRef]
- Ludmerszki, E.; Chounramany, S.; Oláh, C.; Kátay, G.; Rácz, I.; Almási, A.; Solti, Á.; Bélai, I.; Rudnóy, S. Protective role of S-methylmethionine-salicylate in maize plants infected with Maize dwarf mosaic virus. Eur. J. Plant Pathol. 2017, 149, 145–156. [Google Scholar] [CrossRef]
- Ivanov, K.I.; Eskelin, K.; Bašic, M.; De, S.; Lõhmus, A.; Varjosalo, M.; Mäkinen, K. Molecular insights into the function of the viral RNA silencing suppressor HCPro. Plant J. 2016, 85, 30–45. [Google Scholar] [CrossRef] [PubMed]
- De, S.; Chavez-Calvillo, G.; Wahlsten, M.; Mäkinen, K. Disruption of the methionine cycle and reduced cellular gluthathione levels underlie potex–potyvirus synergism in Nicotiana benthamiana. Mol. Plant Pathol. 2018, 19, 1820–1835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, G.; Kong, J.; Cui, D.; Zhao, H.; Niu, Y.; Xu, M.; Jiang, G.; Zhao, Y.; Wang, W. Resistance against Ralstonia solanacearum in tomato depends on the methionine cycle and the γ-aminobutyric acid metabolic pathway. Plant J. 2019, 97, 1032–1047. [Google Scholar] [CrossRef] [PubMed]
- Tirnaz, S.; Batley, J. DNA methylation: Toward crop disease resistance improvement. Trends Plant Sci. 2019, 24, 1137–1150. [Google Scholar] [CrossRef]
- Geng, S.; Kong, X.; Song, G.; Jia, M.; Guan, J.; Wang, F.; Qin, Z.; Wu, L.; Lan, X.; Li, A.; et al. DNA methylation dynamics during the interaction of wheat progenitor Aegilops tauschii with the obligate biotrophic fungus Blumeria graminis f. sp. tritici. New Phytol. 2019, 221, 1023–1035. [Google Scholar] [CrossRef] [Green Version]
- Yu, A.; Lepère, G.; Jay, F.; Wang, J.; Bapaume, L.; Wang, Y.; Abraham, A.L.; Penterman, J.; Fischer, R.L.; Voinnet, O.; et al. Dynamics and biological relevance of DNA demethylation in Arabidopsis antibacterial defense. Proc. Natl. Acad. Sci. USA 2013, 110, 2389–2394. [Google Scholar] [CrossRef] [Green Version]
- Noctor, G.; Queval, G.; Mhamdi, A.; Chaouch, S.; Foyer, C.H. Glutathione. Arab. Book 2011, 9, 1–32. [Google Scholar] [CrossRef]
- Foyer, C.H.; Rennenberg, H. Regulation of glutathione synthesis and its role in abiotic and biotic stress defence. In Sulfur Nutrition and Sulfur Assimilation in Higher Plants; Brunold, C., Rennenberg, H., De Kok, L.J., Stulen, I., Davidian, J.C., Eds.; Paul Haupt: Bern, Switzerland, 2000; pp. 127–153. [Google Scholar]
- Gullner, G.; Kőmíves, T. Defense reactions of infected plants: Roles of glutathione and glutathione S-transferase enzymes. Acta Phytopathol. Entomol. Hung. 2006, 41, 3–10. [Google Scholar] [CrossRef]
- Noctor, G.; Mhamdi, A.; Chaouch, S.; Han, Y.; Neukermans, J.; Marquez-Garcia, B.; Queval, G.; Foyer, C.H. Glutathione in plants: An integrated overview. Plant Cell Environ. 2012, 35, 454–484. [Google Scholar] [CrossRef]
- Gullner, G.; Zechmann, B.; Künstler, A.; Király, L. The signaling roles of glutathione in plant disease resistance. In Glutathione in Plant Growth, Development, and Stress Tolerance; Hossain, M.A., Mostofa, M.G., Vivancos, P.D., Burritt, D.J., Fujita, M., Tran, L.S.P., Eds.; Springer: Cham, Switzerland, 2017; pp. 331–357. ISBN 9783319666822. [Google Scholar]
- Zechmann, B. Subcellular roles of glutathione in mediating plant defense during biotic stress. Plants 2020, 9, 1067. [Google Scholar] [CrossRef]
- Fodor, J.; Gullner, G.; Ádám, A.L.; Barna, B.; Kőmíves, T.; Király, Z. Local and systemic responses of antioxidants to Tobacco mosaic virus infection and to salicylic acid in tobacco: Role in systemic acquired resistance. Plant Physiol. 1997, 114, 1443–1451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vanacker, H.; Foyer, C.H.; Carver, T.L.W. Changes in apoplastic antioxidants induced by powdery mildew attack in oat genotypes with race non-specific resistance. Planta 1999, 208, 444–452. [Google Scholar] [CrossRef]
- Kuźniak, E.; Skłodowska, M. Differential implication of glutathione, glutathione-metabolizing enzymes and ascorbate in tomato resistance to Pseudomonas syringae. J. Phytopathol. 2004, 152, 529–536. [Google Scholar] [CrossRef]
- Großkinsky, D.K.; Koffler, B.E.; Roitsch, T.; Maier, R.; Zechmann, B. Compartment-specific antioxidative defense in Arabidopsis against virulent and avirulent Pseudomonas syringae. Phytopathology 2012, 102, 662–673. [Google Scholar] [CrossRef] [Green Version]
- Harrach, B.D.; Baltruschat, H.; Barna, B.; Fodor, J.; Kogel, K.H. The mutualistic fungus Piriformospora indica protects barley roots from a loss of antioxidant capacity caused by the necrotrophic pathogen Fusarium culmorum. Mol. Plant-Microbe Interact. 2013, 26, 599–605. [Google Scholar] [CrossRef] [Green Version]
- Zechmann, B.; Zellnig, G.; Müller, M. Changes in the subcellular distribution of glutathione during virus infection in Cucurbita pepo (L.). Plant Biol. 2005, 7, 49–57. [Google Scholar] [CrossRef]
- Mukaihara, T.; Hatanaka, T.; Nakano, M.; Oda, K. Ralstonia solanacearum type III effector RipAY is a glutathione-degrading enzyme that is activated by plant cytosolic thioredoxins and suppresses plant immunity. mBio 2016, 7, e00359-16. [Google Scholar] [CrossRef] [Green Version]
- Harrach, B.D.; Fodor, J.; Pogány, M.; Preuss, J.; Barna, B. Antioxidant, ethylene and membrane leakage responses to powdery mildew infection of near-isogenic barley lines with various types of resistance. Eur. J. Plant Pathol. 2008, 121, 21–33. [Google Scholar] [CrossRef]
- Chen, X.; Li, S.; Zhao, X.; Zhu, X.; Wang, Y.; Xuan, Y.; Liu, X.; Fan, H.; Chen, L.; Duan, Y. Modulation of (Homo)glutathione metabolism and H2O2 accumulation during soybean cyst nematode infections in susceptible and resistant soybean cultivars. Int. J. Mol. Sci. 2020, 21, 388. [Google Scholar] [CrossRef] [Green Version]
- Ghanta, S.; Bhattacharyya, D.; Sinha, R.; Banerjee, A.; Chattopadhyay, S. Nicotiana tabacum overexpressing γ-ECS exhibits biotic stress tolerance likely through NPR1-dependent salicylic acid-mediated pathway. Planta 2011, 233, 895–910. [Google Scholar] [CrossRef]
- Ghanta, S.; Datta, R.; Bhattacharyya, D.; Sinha, R.; Kumar, D.; Hazra, S.; Mazumdar, A.B.; Chattopadhyay, S. Multistep involvement of glutathione with salicylic acid and ethylene to combat environmental stress. J. Plant Physiol. 2014, 171, 940–950. [Google Scholar] [CrossRef] [PubMed]
- Künstler, A.; Király, L.; Kátay, G.; Enyedi, A.J.; Gullner, G. Glutathione can oompensate for salicylic acid deficiency in tobacco to maintain resistance to Tobacco mosaic virus. Front. Plant Sci. 2019, 10, 1115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomez, L.D.; Noctor, G.; Knight, M.R.; Foyer, C.H. Regulation of calcium signalling and gene expression by glutathione. J. Exp. Bot. 2004, 55, 1851–1859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hausladen, A.; Kunert, K.J. Effects of artificially enhanced levels of ascorbate and glutathione on the enzymes monodehydroascorbate reductase, dehydroascorbate reductase, and glutathione reductase in spinach (Spinacia oleracea). Physiol. Plant. 1990, 79, 384–388. [Google Scholar] [CrossRef]
- Farago, S.; Brunold, C. Regulation of thiol contents in maize roots by intermediates and effectors of glutathione synthesis. J. Plant Physiol. 1994, 144, 433–437. [Google Scholar] [CrossRef]
- Gullner, G.; Tóbiás, I.; Fodor, J.; Kőmíves, T. Elevation of glutathione level and activation of glutathione-related enzymes affect virus infection in tobacco. Free Radic. Res. 1999, 31, 155–161. [Google Scholar] [CrossRef]
- Zechmann, B.; Zellnig, G.; Urbanek-Krajnc, A.; Müller, M. Artificial elevation of glutathione affects symptom development in ZYMV-infected Cucurbita pepo L. plants. Arch. Virol. 2007, 152, 747–762. [Google Scholar] [CrossRef]
- Clemente-Moreno, M.J.; Díaz-Vivancos, P.; Barba-Espín, G.; Hernández, J.A. Benzothiadiazole and l-2-oxothiazolidine-4-carboxylic acid reduce the severity of Sharka symptoms in pea leaves: Effect on antioxidative metabolism at the subcellular level. Plant Biol. 2010, 12, 88–97. [Google Scholar] [CrossRef]
- Clemente-Moreno, M.J.; Díaz-Vivancos, P.; Piqueras, A.; Hernández, J.A. Plant growth stimulation in Prunus species plantlets by BTH or OTC treatments under in vitro conditions. J. Plant Physiol. 2012, 169, 1074–1083. [Google Scholar] [CrossRef]
- Clemente-Moreno, M.J.; Díaz-Vivancos, P.; Rubio, M.; Fernández-García, N.; Hernández, J.A. Chloroplast protection in Plum pox virus-infected peach plants by L-2-oxo-4-thiazolidine-carboxylic acid treatments: Effect in the proteome. Plant Cell Environ. 2013, 36, 640–654. [Google Scholar] [CrossRef] [Green Version]
- Künstler, A.; Kátay, G.; Gullner, G.; Király, L. Artificial elevation of glutathione contents in salicylic acid-deficient tobacco (Nicotiana tabacum cv. Xanthi NahG) reduces susceptibility to the powdery mildew pathogen Euoidium longipes. Plant Biol. 2020, 22, 70–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Höller, K.; Király, L.; Künstler, A.; Müller, M.; Gullner, G.; Fattinger, M.; Zechmann, B. Enhanced glutathione metabolism is correlated with sulfur-induced resistance in Tobacco mosaic virus-infected genetically susceptible Nicotiana tabacum plants. Mol. Plant-Microbe Interact. 2010, 23, 1448–1459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Király, L.; Künstler, A.; Höller, K.; Fattinger, M.; Juhász, C.; Müller, M.; Gullner, G.; Zechmann, B. Sulfate supply influences compartment specific glutathione metabolism and confers enhanced resistance to Tobacco mosaic virus during a hypersensitive response. Plant Physiol. Biochem. 2012, 59, 44–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parisy, V.; Poinssot, B.; Owsianowski, L.; Buchala, A.; Glazebrook, J.; Mauch, F. Identification of PAD2 as a γ-glutamylcysteine synthetase highlights the importance of glutathione in disease resistance of Arabidopsis. Plant J. 2007, 49, 159–172. [Google Scholar] [CrossRef]
- Datta, R.; Chattopadhyay, S. Glutathione as a crucial modulator of phytohormone signalling during pathogen defence in plants. Proc. Indian Natl. Sci. Acad. 2018, 84, 581–597. [Google Scholar] [CrossRef]
- Kumar, D. Salicylic acid signaling in disease resistance. Plant Sci. 2014, 228, 127–134. [Google Scholar] [CrossRef]
- Doke, N. Involvement of superoxide anion generation in the hypersensitive response of potato tuber tissues to infection with an incompatible race of Phytophthora infestans and to the hyphal wall components. Physiol. Plant Pathol. 1983, 23, 345–357. [Google Scholar] [CrossRef]
- Levine, A.; Tenhaken, R.; Dixon, R.; Lamb, C. H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell 1994, 79, 583–593. [Google Scholar] [CrossRef]
- Delledonne, M.; Zeier, J.; Marocco, A.; Lamb, C. Signal interactions between nitric oxide and reactive oxygen intermediates in the plant hypersensitive disease resistance response. Proc. Natl. Acad. Sci. USA 2001, 98, 13454–13459. [Google Scholar] [CrossRef] [Green Version]
- Torres, M.A.; Jones, J.D.G.; Dangl, J.L. Reactive oxygen species signaling in response to pathogens. Plant Physiol. 2006, 141, 373–378. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Xu, X.; Leng, X.; He, M.; Wang, J.; Cheng, S.; Wu, H. Roles of reactive oxygen species in cell signaling pathways and immune responses to viral infections. Arch. Virol. 2017, 162, 603–610. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Chaouch, S.; Mhamdi, A.; Queval, G.; Zechmann, B.; Noctor, G. Functional analysis of Arabidopsis mutants points to novel roles for glutathione in coupling H2O2 to activation of salicylic acid accumulation and signaling. Antioxid. Redox Signal. 2013, 18, 2106–2121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lindermayr, C. Crosstalk between reactive oxygen species and nitric oxide in plants: Key role of S-nitrosoglutathione reductase. Free Radic. Biol. Med. 2018, 122, 110–115. [Google Scholar] [CrossRef] [PubMed]
- Kovacs, I.; Durner, J.; Lindermayr, C. Crosstalk between nitric oxide and glutathione is required for nonexpressor of pathogenesis-related GENES 1 (NPR1)-dependent defense signaling in Arabidopsis thaliana. New Phytol. 2015, 208, 860–872. [Google Scholar] [CrossRef]
- Feechan, A.; Kwon, E.; Yun, B.W.; Wang, Y.; Pallas, J.A.; Loake, G.J. A central role for S-nitrosothiols in plant disease resistance. Proc. Natl. Acad. Sci. USA 2005, 102, 8054–8059. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.; Ma, M.; Chen, T.; Zhang, L.; Fan, L.; Zhang, W.; Wei, B.; Li, S.; Xuan, W.; Noctor, G.; et al. Glutathione-dependent denitrosation of GSNOR1 promotes oxidative signalling downstream of H2O2. Plant Cell Environ. 2020, 43, 1175–1191. [Google Scholar] [CrossRef]
- Leterrier, M.; Chaki, M.; Airaki, M.; Valderrama, R.; Palma, J.M.; Barroso, J.B.; Corpas, F.J. Function of S-nitrosoglutathione reductase (GSNOR) in plant development and under biotic/abiotic stress. Plant Signal. Behav. 2011, 6, 789–793. [Google Scholar] [CrossRef] [Green Version]
- Mou, Z.; Fan, W.; Dong, X. Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 2003, 113, 935–944. [Google Scholar] [CrossRef] [Green Version]
- Tada, Y.; Spoel, S.H.; Pajerowska-Mukhtar, K.; Mou, Z.; Song, J.; Wang, C.; Zuo, J.; Dong, X. Plant immunity requires conformational charges of NPR1 via S-nitrosylation and thioredoxins. Science 2008, 321, 952–956. [Google Scholar] [CrossRef] [Green Version]
- Mata-Pérez, C.; Spoel, S.H. Thioredoxin-mediated redox signalling in plant immunity. Plant Sci. 2019, 279, 27–33. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, D.; Chu, J.Y.; Boyle, P.; Wang, Y.; Brindle, I.D.; De Luca, V.; Després, C. The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Rep. 2012, 1, 639–647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lindermayr, C.; Sell, S.; Müller, B.; Leister, D.; Durner, J. Redox regulation of the NPR1-TGA1 system of Arabidopsis thaliana by nitric oxide. Plant Cell 2010, 22, 2894–2907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaffney, T.; Friedrich, L.; Vernooij, B.; Negrotto, D.; Nye, G.; Uknes, S.; Ward, E.; Kessmann, H.; Ryals, J. Requirement of salicylic acid for the induction of systemic acquired resistance. Science 1993, 261, 754–756. [Google Scholar] [CrossRef] [PubMed]
- Costa, D.M.A.; Gómez, S.V.; de Araújo, S.S.; Pereira, M.S.; Alves, R.B.; Favaro, D.C.; Hengge, A.C.; Nagem, R.A.P.; Brandão, T.A.S. Catalytic mechanism for the conversion of salicylate into catechol by the flavin-dependent monooxygenase salicylate hydroxylase. Int. J. Biol. Macromol. 2019, 129, 588–600. [Google Scholar] [CrossRef] [Green Version]
- Delaney, T.P.; Uknes, S.; Vernooij, B.; Friedrich, L.; Weymann, K.; Negrotto, D.; Gaffney, T.; Gut-Rella, M.; Kessmann, H.; Ward, E.; et al. A central role of salicylic acid in plant disease resistance. Science 1994, 266, 1247–1250. [Google Scholar] [CrossRef] [Green Version]
- Pandey, D.; Rajendran, S.R.C.K.; Gaur, M.; Sajeesh, P.K.; Kumar, A. Plant defense signaling and responses against necrotrophic fungal pathogens. J. Plant Growth Regul. 2016, 35, 1159–1174. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, F.; Melotto, M.; Yao, J.; He, S.Y. Jasmonate signaling and manipulation by pathogens and insects. J. Exp. Bot. 2017, 68, 1371–1385. [Google Scholar] [CrossRef]
- De Vleesschauwer, D.; Gheysen, G.; Höfte, M. Hormone defense networking in rice: Tales from a different world. Trends Plant Sci. 2013, 18, 555–565. [Google Scholar] [CrossRef]
- Han, Y.; Mhamdi, A.; Chaouch, S.; Noctor, G. Regulation of basal and oxidative stress-triggered jasmonic acid-related gene expression by glutathione. Plant Cell Environ. 2013, 36, 1135–1146. [Google Scholar] [CrossRef]
- Caarls, L.; Pieterse, C.M.J.; Van Wees, S.C.M. How salicylic acid takes transcriptional control over jasmonic acid signaling. Front. Plant Sci. 2015, 6, 170. [Google Scholar] [CrossRef]
- Koornneef, A.; Leon-Reyes, A.; Ritsema, T.; Verhage, A.; Den Otter, F.C.; Van Loon, L.C.; Pieterse, C.M.J. Kinetics of salicylate-mediated suppression of jasmonate signaling reveal a role for redox modulation. Plant Physiol. 2008, 147, 1358–1368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sauter, M.; Moffatt, B.; Saechao, M.C.; Hell, R.; Wirtz, M. Methionine salvage and S-adenosylmethionine: Essential links between sulfur, ethylene and polyamine biosynthesis. Biochem. J. 2013, 451, 145–154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stasolla, C. Glutathione redox regulation of in vitro embryogenesis. Plant Physiol. Biochem. 2010, 48, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Datta, R.; Kumar, D.; Sultana, A.; Hazra, S.; Bhattacharyya, D.; Chattopadhyay, S. Glutathione regulates 1-aminocyclopropane-1-carboxylate synthase transcription via WRKY33 and 1-aminocyclopropane-1-carboxylate oxidase by modulating messenger RNA stability to induce ethylene synthesis during stress. Plant Physiol. 2015, 169, 2963–2981. [Google Scholar] [CrossRef] [PubMed]
- Gullner, G.; Kőmíves, T.; Király, L.; Schröder, P. Glutathione S-transferase enzymes in plant-pathogen interactions. Front. Plant Sci. 2018, 9, 1836. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pei, D.; Ma, H.; Zhang, Y.; Ma, Y.; Wang, W.; Geng, H.; Wu, J.; Li, C. Virus-induced gene silencing of a putative glutathione S-transferase gene compromised Ol-1-mediated resistance against powdery mildew in tomato. Plant Mol. Biol. Rep. 2011, 29, 972–978. [Google Scholar] [CrossRef]
- Peters, L.P.; Carvalho, G.; Vilhena, M.B.; Creste, S.; Azevedo, R.A.; Monteiro-Vitorello, C.B. Functional analysis of oxidative burst in sugarcane smut-resistant and -susceptible genotypes. Planta 2017, 245, 749–764. [Google Scholar] [CrossRef] [Green Version]
- Gong, Q.; Yang, Z.; Chen, E.; Sun, G.; He, S.; Butt, H.I.; Zhang, C.; Zhang, X.; Yang, Z.; Du, X.; et al. A Phi-class glutathione S-transferase gene for Verticillium wilt resistance in Gossypium arboreum identified in a genome-wide association study. Plant Cell Physiol. 2018, 59, 275–289. [Google Scholar] [CrossRef]
- Czerniawski, P.; Bednarek, P. Glutathione S-transferases in the biosynthesis of sulfur-containing secondary metabolites in Brassicaceae plants. Front. Plant Sci. 2018, 9, 871. [Google Scholar] [CrossRef]
- Piślewska-Bednarek, M.; Nakano, R.T.; Hiruma, K.; Pastorczyk, M.; Sanchez-Vallet, A.; Singkaravanit-Ogawa, S.; Ciesiołka, D.; Takano, Y.; Molina, A.; Schulze-Lefert, P.; et al. Glutathione transferase U13 functions in pathogen-triggered glucosinolate metabolism. Plant Physiol. 2018, 176, 538–551. [Google Scholar] [CrossRef] [Green Version]
- Van Loon, L.C.; Rep, M.; Pieterse, C.M.J. Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 2006, 44, 135–162. [Google Scholar] [CrossRef] [Green Version]
- Stec, B. Plant thionins—The structural perspective. Cell. Mol. Life Sci. 2006, 63, 1370–1385. [Google Scholar] [CrossRef] [PubMed]
- Silverstein, K.A.T.; Moskal, W.A.; Wu, H.C.; Underwood, B.A.; Graham, M.A.; Town, C.D.; VandenBosch, K.A. Small cysteine-rich peptides resembling antimicrobial peptides have been under-predicted in plants. Plant J. 2007, 51, 262–280. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.H.; Ng, T.B. Sesquin, a potent defensin-like antimicrobial peptide from ground beans with inhibitory activities toward tumor cells and HIV-1 reverse transcriptase. Peptides 2005, 26, 1120–1126. [Google Scholar] [CrossRef] [PubMed]
- Lacerda, A.F.; Vasconcelos, É.A.R.; Pelegrini, P.B.; Grossi-de-Sa, M.F. Antifungal defensins and their role in plant defense. Front. Microbiol. 2014, 5, 116. [Google Scholar] [CrossRef] [Green Version]
- Sathoff, A.E.; Samac, D.A. Antibacterial activity of plant defensins. Mol. Plant-Microbe Interact. 2019, 32, 507–514. [Google Scholar] [CrossRef]
- Cools, T.L.; Struyfs, C.; Cammue, B.P.; Thevissen, K. Antifungal plant defensins: Increased insight in their mode of action as a basis for their use to combat fungal infections. Future Microbiol. 2017, 12, 441–454. [Google Scholar] [CrossRef]
- Wilmes, M.; Cammue, B.P.A.; Sahl, H.G.; Thevissen, K. Antibiotic activities of host defense peptides: More to it than lipid bilayer perturbation. Nat. Prod. Rep. 2011, 28, 1350–1358. [Google Scholar] [CrossRef]
- Baxter, A.A.; Richter, V.; Lay, F.T.; Poon, I.K.H.; Adda, C.G.; Veneer, P.K.; Phan, T.K.; Bleackley, M.R.; Anderson, M.A.; Kvansakul, M.; et al. The tomato defensin TPP3 binds phosphatidylinositol (4,5)-bisphosphate via a conserved dimeric cationic grip conformation to mediate cell lysis. Mol. Cell. Biol. 2015, 35, 1964–1978. [Google Scholar] [CrossRef] [Green Version]
- Oard, S.V. Deciphering a mechanism of membrane permeabilization by α-hordothionin peptide. Biochim. Biophys. Acta Biomembr. 2011, 1808, 1737–1745. [Google Scholar] [CrossRef] [Green Version]
- Vieira, M.E.B.; Vasconcelos, I.M.; Machado, O.L.T.; Gomes, V.M.; De Oliveira Carvalho, A. Isolation, characterization and mechanism of action of an antimicrobial peptide from Lecythis pisonis seeds with inhibitory activity against Candida albicans. Acta Biochim. Biophys. Sin. (Shanghai) 2015, 47, 716–729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aerts, A.M.; Carmona-Gutierrez, D.; Lefevre, S.; Govaert, G.; François, I.E.J.A.; Madeo, F.; Santos, R.; Cammue, B.P.A.; Thevissen, K. The antifungal plant defensin RsAFP2 from radish induces apoptosis in a metacaspase independent way in Candida albicans. FEBS Lett. 2009, 583, 2513–2516. [Google Scholar] [CrossRef] [PubMed]
- Penninckx, I.A.M.A.; Eggermont, K.; Terras, F.R.G.; Thomma, B.P.H.J.; De Samblanx, G.W.; Buchala, A.; Métraux, J.P.; Manners, J.M.; Broekaert, W.F. Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid-independent pathway. Plant Cell 1996, 8, 2309–2323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hiruma, K.; Nishiuchi, T.; Kato, T.; Bednarek, P.; Okuno, T.; Schulze-Lefert, P.; Takano, Y. Arabidopsis enhanced disease RESISTANCE 1 is required for pathogen-induced expression of plant defensins in nonhost resistance, and acts through interference of MYC2-mediated repressor function. Plant J. 2011, 67, 980–992. [Google Scholar] [CrossRef] [PubMed]
- Lacerda, A.F.; Del Sarto, R.P.; Silva, M.S.; de Vasconcelos, E.A.R.; Coelho, R.R.; dos Santos, V.O.; Godoy, C.V.; Seixas, C.D.S.; da Silva, M.C.M.; Grossi-de-Sa, M.F. The recombinant pea defensin Drr230a is active against impacting soybean and cotton pathogenic fungi from the genera Fusarium, Colletotrichum and Phakopsora. 3 Biotech 2016, 6, 59. [Google Scholar] [CrossRef] [Green Version]
- Velivelli, S.L.S.; Islam, K.T.; Hobson, E.; Shah, D.M. Modes of action of a Bi-domain plant defensin MtDef5 against a bacterial pathogen Xanthomonas campestris. Front. Microbiol. 2018, 9, 934. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Ganai, B.A.; Kamili, A.N.; Bhat, A.A.; Mir, Z.A.; Bhat, J.A.; Tyagi, A.; Islam, S.T.; Mushtaq, M.; Yadav, P.; et al. Pathogenesis-related proteins and peptides as promising tools for engineering plants with multiple stress tolerance. Microbiol. Res. 2018, 212–213, 29–37. [Google Scholar] [CrossRef]
- Ghag, S.B.; Shekhawat, U.K.S.; Ganapathi, T.R. Petunia floral defensins with unique prodomains as novel candidates for development of Fusarium wilt resistance in transgenic banana plants. PLoS ONE 2012, 7, e39557. [Google Scholar] [CrossRef]
- Asano, T.; Miwa, A.; Maeda, K.; Kimura, M.; Nishiuchi, T. The secreted antifungal protein thionin 2.4 in Arabidopsis thaliana suppresses the toxicity of a fungal fruit body lectin from Fusarium graminearum. PLoS Pathog. 2013, 9, e1003581. [Google Scholar] [CrossRef]
- Sasaki, K.; Kuwabara, C.; Umeki, N.; Fujioka, M.; Saburi, W.; Matsui, H.; Abe, F.; Imai, R. The cold-induced defensin TAD1 confers resistance against snow mold and Fusarium head blight in transgenic wheat. J. Biotechnol. 2016, 228, 3–7. [Google Scholar] [CrossRef]
- Hao, G.; Bakker, M.G.; Kim, H.S. Enhanced resistance to Fusarium graminearum in transgenic Arabidopsis plants expressing a modified plant thionin. Phytopathology 2020, 110, 1056–1066. [Google Scholar] [CrossRef] [PubMed]
- Mansfield, J.W. Antimicrobial compounds and resistance. In Mechanisms of Resistance to Plant Diseases; Slusarenko, A.J., Fraser, R.S.S., van Loon, L.C., Eds.; Springer: Dordrecht, The Netherlands, 2000; pp. 325–370. [Google Scholar]
- Nwachukwu, I.D.; Slusarenko, A.J.; Gruhlke, M.C.H. Sulfur and sulfur compounds in plant defence. Nat. Prod. Commun. 2012, 7. [Google Scholar] [CrossRef] [Green Version]
- Ahuja, I.; Kissen, R.; Bones, A.M. Phytoalexins in defense against pathogens. Trends Plant Sci. 2012, 17, 73–90. [Google Scholar] [CrossRef] [PubMed]
- Pedras, M.S.C.; Yaya, E.E.; Glawischnig, E. The phytoalexins from cultivated and wild crucifers: Chemistry and biology. Nat. Prod. Rep. 2011, 28, 1381–1405. [Google Scholar] [CrossRef]
- Bednarek, P. Sulfur-containing secondary metabolites from Arabidopsis thaliana and other Brassicaceae with function in plant immunity. ChemBioChem 2012, 13, 1846–1859. [Google Scholar] [CrossRef]
- Pedras, M.S.C.; Abdoli, A. Pathogen inactivation of cruciferous phytoalexins: Detoxification reactions, enzymes and inhibitors. RSC Adv. 2017, 7, 23633–23646. [Google Scholar] [CrossRef] [Green Version]
- Falk, K.L.; Tokuhisa, J.G.; Gershenzon, J. The effect of sulfur nutrition on plant glucosinolate content: Physiology and molecular mechanisms. Plant Biol. 2007, 9, 573–581. [Google Scholar] [CrossRef]
- Maruyama-Nakashita, A.; Ohkama-Ohtsu, N. Sulfur assimilation and glutathione metabolism in plants. In Glutathione in Plant Growth, Development, and Stress Tolerance; Hossain, M.A., Mostofa, M.G., Vivancos, P.D., Burritt, D.J., Fujita, M., Tran, L.S.P., Eds.; Springer: Cham, Switzerland, 2017; pp. 287–308. ISBN 9783319666822. [Google Scholar]
- Klein, A.P.; Anarat-Cappillino, G.; Sattely, E.S. Minimum set of cytochromes P450 for reconstituting the biosynthesis of camalexin, a major Arabidopsis antibiotic. Angew. Chemie Int. Ed. 2013, 52, 13625–13628. [Google Scholar] [CrossRef] [Green Version]
- Su, T.; Xu, J.; Li, Y.; Lei, L.; Zhao, L.; Yang, H.; Feng, J.; Liu, G.; Rena, D. Glutathione-indole-3-acetonitrile is required for camalexin biosynthesis in Arabidopsis thaliana. Plant Cell 2011, 23, 364–380. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.Y.; Liu, X.T.; Chen, Y.; Xu, X.J.; Yu, B.; Zhang, S.Q.; Li, Q.; He, Z.H. Arabidopsis acetyl-amido synthetase GH3.5 involvement in camalexin biosynthesis through conjugation of indole-3-carboxylic acid and cysteine and upregulation of camalexin biosynthesis genes. J. Integr. Plant Biol. 2012, 54, 471–485. [Google Scholar] [CrossRef]
- Schuhegger, R.; Rauhut, T.; Glawischnig, E. Regulatory variability of camalexin biosynthesis. J. Plant Physiol. 2007, 164, 636–644. [Google Scholar] [CrossRef] [PubMed]
- Böttcher, C.; Westphal, L.; Schmotz, C.; Prade, E.; Scheel, D.; Glawischnig, E. The multifunctional enzyme CYP71B15 (PHYTOALEXIN DEFICIENT3) converts cysteine-indole-3-acetonitrile to camalexin in the indole-3-acetonitrile metabolic network of Arabidopsis thaliana. Plant Cell 2009, 21, 1830–1845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomma, B.P.H.J.; Nelissen, I.; Eggermont, K.; Broekaert, W.F. Deficiency in phytoalexin production causes enhanced susceptibility of Arabidopsis thaliana to the fungus Alternaria brassicicola. Plant J. 1999, 19, 163–171. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, S.; Plotnikova, J.M.; De Lorenzo, G.; Ausubel, F.M. Arabidopsis local resistance to Botrytis cinerea involves salicylic acid and camalexin and requires EDS4 and PAD2, but not SID2, EDS5 or PAD4. Plant J. 2003, 35, 193–205. [Google Scholar] [CrossRef] [Green Version]
- Bohman, S.; Staal, J.; Thomma, B.P.H.J.; Wang, M.; Dixelius, C. Characterisation of an Arabidopsis-Leptosphaeria maculans pathosystem: Resistance partially requires camalexin biosynthesis and is independent of salicylic acid, ethylene and jasmonic acid signalling. Plant J. 2004, 37, 9–20. [Google Scholar] [CrossRef]
- Liu, S.; Bartnikas, L.M.; Volko, S.M.; Ausubel, F.M.; Tang, D. Mutation of the glucosinolate biosynthesis enzyme cytochrome P450 83A1 monooxygenase increases camalexin accumulation and powdery mildew resistance. Front. Plant Sci. 2016, 7, 227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nafisi, M.; Goregaoker, S.; Botanga, C.J.; Glawischnig, E.; Olsen, C.E.; Halkier, B.A.; Glazebrook, J. Arabidopsis cytochrome P450 monooxygenase 71A13 catalyzes the conversion of indole-3-acetaldoxime in camalexin synthesis. Plant Cell 2007, 19, 2039–2052. [Google Scholar] [CrossRef] [Green Version]
- Ren, D.; Liu, Y.; Yang, K.Y.; Han, L.; Mao, G.; Glazebrook, J.; Zhang, S. A fungal-responsive MAPK cascade regulates phytoalexin biosynthesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2008, 105, 5638–5643. [Google Scholar] [CrossRef] [Green Version]
- Mao, G.; Meng, X.; Liu, Y.; Zheng, Z.; Chen, Z.; Zhang, S. Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 2011, 23, 1639–1653. [Google Scholar] [CrossRef] [Green Version]
- Khare, D.; Choi, H.; Huh, S.U.; Bassin, B.; Kim, J.; Martinoia, E.; Sohn, K.H.; Paek, K.H.; Lee, Y.; Chrispeels, M.J. Arabidopsis ABCG34 contributes to defense against necrotrophic pathogens by mediating the secretion of camalexin. Proc. Natl. Acad. Sci. USA 2017, 114, E5712–E5720. [Google Scholar] [CrossRef] [Green Version]
- Resende, M.L.V.; Flood, J.; Ramsden, J.D.; Rowan, M.G.; Beale, M.H.; Cooper, R.M. Novel phytoalexins including elemental sulphur in the resistance of cocoa (Theobroma cacao L.) to Verticillium wilt (Verticillium dahliae Kleb.). Physiol. Mol. Plant Pathol. 1996, 48, 347–359. [Google Scholar] [CrossRef]
- Williams, J.S.; Hall, S.A.; Hawkesford, M.J.; Beale, M.H.; Cooper, R.M. Elemental sulfur and thiol accumulation in tomato and defense against a fungal vascular pathogen. Plant Physiol. 2002, 128, 150–159. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.S.; Cooper, R.M. Elemental sulphur is produced by diverse plant families as a component of defence against fungal and bacterial pathogens. Physiol. Mol. Plant Pathol. 2003, 63, 3–16. [Google Scholar] [CrossRef]
- Cooper, R.M.; Williams, J.S. Elemental sulphur as an induced antifungal substance in plant defence. J. Exp. Bot. 2004, 55, 1947–1953. [Google Scholar] [CrossRef] [PubMed]
- Rahimi, F.; Rahmanpour, S. Overcoming glucosinolate-myrosinase-isothiocyanate defense system by plant pathogenic fungi. Int. J. Second. Metab. 2020, 7, 19–27. [Google Scholar] [CrossRef]
- Halkier, B.A.; Gershenzon, J. Biology and biochemistry of glucosinolates. Annu. Rev. Plant Biol. 2006, 57, 303–333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aghajanzadeh, T.; Hawkesford, M.J.; De Kok, L.J. The significance of glucosinolates for sulfur storage in Brassicaceae seedlings. Front. Plant Sci. 2014, 5, 5. [Google Scholar] [CrossRef] [Green Version]
- Koroleva, O.A.; Cramer, R. Single-cell proteomic analysis of glucosinolate-rich S-cells in Arabidopsis thaliana. Methods 2011, 54, 413–423. [Google Scholar] [CrossRef]
- Nour-Eldin, H.H.; Andersen, T.G.; Burow, M.; Madsen, S.R.; Jørgensen, M.E.; Olsen, C.E.; Dreyer, I.; Hedrich, R.; Geiger, D.; Halkier, B.A. NRT/PTR transporters are essential for translocation of glucosinolate defence compounds to seeds. Nature 2012, 488, 531–534. [Google Scholar] [CrossRef]
- Xu, J.; Meng, J.; Meng, X.; Zhao, Y.; Liu, J.; Sun, T.; Liu, Y.; Wang, Q.; Zhang, S. Pathogen-responsive MPK3 and MPK6 reprogram the biosynthesis of indole glucosinolates and their derivatives in Arabidopsis immunity. Plant Cell 2016, 28, 1144–1162. [Google Scholar] [CrossRef] [Green Version]
- Sugiyama, R.; Hirai, M.Y. Atypical myrosinase as a mediator of glucosinolate functions in plants. Front. Plant Sci. 2019, 10, 1008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wittstock, U.; Meier, K.; Dörr, F.; Ravindran, B.M. NSP-dependent simple nitrile formation dominates upon breakdown of major aliphatic glucosinolates in roots, seeds, and seedlings of Arabidopsis thaliana Columbia-0. Front. Plant Sci. 2016, 7, 1821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bednarek, P.; Piślewska-Bednarek, M.; Svatoš, A.; Schneider, B.; Doubský, J.; Mansurova, M.; Humphry, M.; Consonni, C.; Panstruga, R.; Sanchez-Vallet, A.; et al. A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 2009, 323, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Bednarek, P.; Piślewska-Bednarek, M.; Ver Loren van Themaat, E.; Maddula, R.K.; Svatoš, A.; Schulze-Lefert, P. Conservation and clade-specific diversification of pathogen-inducible tryptophan and indole glucosinolate metabolism in Arabidopsis thaliana relatives. New Phytol. 2011, 192, 713–726. [Google Scholar] [CrossRef]
- Lu, X.; Dittgen, J.; Pislewska-Bednarek, M.; Molina, A.; Schneider, B.; Svatos, A.; Doubsky, J.; Schneeberger, K.; Weigel, D.; Bednarek, P.; et al. Mutant allele-specific uncoupling of penetration3 functions reveals engagement of the ATP-binding cassette transporter in distinct tryptophan metabolic pathways. Plant Physiol. 2015, 168, 814–827. [Google Scholar] [CrossRef]
- He, Y.; Xu, J.; Wang, X.; He, X.; Wang, Y.; Zhou, J.; Zhang, S.; Meng, X. The Arabidopsis pleiotropic drug resistance transporters PEN3 and PDR12 mediate camalexin secretion for resistance to Botrytis cinerea. Plant Cell 2019, 31, 2206–2222. [Google Scholar] [CrossRef]
- Lipka, V.; Dittgen, J.; Bednarek, P.; Bhat, R.; Wiermer, M.; Stein, M.; Landtag, J.; Brandt, W.; Rosahl, S.; Scheel, D.; et al. Plant science: Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science 2005, 310, 1180–1183. [Google Scholar] [CrossRef] [Green Version]
- Collins, N.C.; Thordal-Christensen, H.; Lipka, V.; Bau, S.; Kombrink, E.; Qiu, J.-L.; Hückelhoven, R.; Stein, M.; Freialdenhoven, A.; Somerville, S.C.; et al. SNARE-protein-mediated disease resistance at the plant cell wall. Nature 2003, 425, 973–977. [Google Scholar] [CrossRef]
- Mellersh, D.G.; Foulds, I.V.; Higgins, V.J.; Heath, M.C. H2O2 plays different roles in determining penetration failure in three diverse plant-fungal interactions. Plant J. 2002, 29, 257–268. [Google Scholar] [CrossRef]
- Schlaeppi, K.; Abou-Mansour, E.; Buchala, A.; Mauch, F. Disease resistance of Arabidopsis to Phytophthora brassicae is established by the sequential action of indole glucosinolates and camalexin. Plant J. 2010, 62, 840–851. [Google Scholar] [CrossRef] [Green Version]
- Zhang, K.; Su, H.; Zhou, J.; Liang, W.; Liu, D.; Li, J. Overexpressing the myrosinase gene TGG1 enhances stomatal defense against Pseudomonas syringae and delays flowering in Arabidopsis. Front. Plant Sci. 2019, 10, 1230. [Google Scholar] [CrossRef] [PubMed]
- Rupp, S.I.; Hornbacher, J.; Horst-Niessen, I.; Schaarschmidt, F.; Riemenschneider, A.; Papenbrock, J. The diurnal rhythm of Brassica napus L. influences contents of sulfur-containing defense compounds and occurrence of vascular occlusions during an infection with Verticillium longisporum. Agronomy 2020, 10, 1227. [Google Scholar] [CrossRef]
- Borlinghaus, J.; Albrecht, F.; Gruhlke, M.; Nwachukwu, I.; Slusarenko, A. Allicin: Chemistry and biological properties. Molecules 2014, 19, 12591–12618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshimoto, N.; Saito, K. S-Alk(en)ylcysteine sulfoxides in the genus Allium: Proposed biosynthesis, chemical conversion, and bioactivities. J. Exp. Bot. 2019, 70, 4123–4137. [Google Scholar] [CrossRef]
- Zhu, G.; Luo, Y.; Xue, M.; Zhou, F.; Zhao, H.; Ji, G.; Liu, F. Resistance of garlic cultivars to Bradysia odoriphaga and its correlation with garlic thiosulfinates. Sci. Rep. 2017, 7, 3249. [Google Scholar] [CrossRef]
- Stice, S.P.; Thao, K.K.; Khang, C.H.; Baltrus, D.A.; Dutta, B.; Kvitko, B.H. Thiosulfinate tolerance is a virulence strategy of an atypical bacterial pathogen of onion. Curr. Biol. 2020, 30, 3130–3140.e6. [Google Scholar] [CrossRef]
- Borlinghaus, J.; Bolger, A.; Schier, C.; Vogel, A.; Usadel, B.; Gruhlke, M.C.; Slusarenko, A.J. Genetic and molecular characterization of multicomponent resistance of Pseudomonas against allicin. Life Sci. Alliance 2020, 3, e202000670. [Google Scholar] [CrossRef] [Green Version]
- Kyung, K.H.; Fleming, H.P. S-methyl-l-cysteine sulfoxide as the precursor of methyl methanethiolsulfinate, the principal antibacterial compound in cabbage. J. Food Sci. 1994, 59, 350–355. [Google Scholar] [CrossRef]
- Kyung, K.H.; Fleming, H.P. Antimicrobial activity-of sulfur compounds derived from cabbage. J. Food Prot. 1997, 60, 67–71. [Google Scholar] [CrossRef]
- Gruhlke, M.C.H.; Slusarenko, A.J. The biology of reactive sulfur species (RSS). Plant Physiol. Biochem. 2012, 59, 98–107. [Google Scholar] [CrossRef]
- Giles, G.I.; Nasim, M.J.; Ali, W.; Jacob, C. The reactive sulfur species concept: 15 years on. Antioxidants 2017, 6, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corpas, F.J.; González-Gordo, S.; Palma, J.M. Plant peroxisomes: A factory of reactive species. Front. Plant Sci. 2020, 11, 853. [Google Scholar] [CrossRef] [PubMed]
- Gao, R.; Ng, F.K.L.; Liu, P.; Wong, S.M. Hibiscus chlorotic ringspot virus coat protein upregulates sulfur metabolism genes for enhanced pathogen defense. Mol. Plant-Microbe Interact. 2012, 25, 1574–1583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giraud, E.; Ivanova, A.; Gordon, C.S.; Whelan, J.; Considine, M.J. Sulphur dioxide evokes a large scale reprogramming of the grape berry transcriptome associated with oxidative signalling and biotic defence responses. Plant Cell Environ. 2012, 35, 405–417. [Google Scholar] [CrossRef]
- Chen, J.; Liu, T.W.; Hu, W.J.; Simon, M.; Wang, W.H.; Liu, X.; Zheng, H.L. Comparative proteomic analysis of differentially expressed proteins induced by hydrogen sulfide in Spinacia oleracea leaves. PLoS ONE 2014, 9, e105400. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Fang, H.; Pei, Y.; Jin, Z.; Zhang, L.; Liu, D. WRKY transcription factors down-regulate the expression of H2S-generating genes, LCD and DES in Arabidopsis thaliana. Sci. Bull. 2015, 60, 995–1001. [Google Scholar] [CrossRef] [Green Version]
- Shi, H.; Ye, T.; Han, N.; Bian, H.; Liu, X.; Chan, Z. Hydrogen sulfide regulates abiotic stress tolerance and biotic stress resistance in Arabidopsis. J. Integr. Plant Biol. 2015, 57, 628–640. [Google Scholar] [CrossRef]
- Abe, K.; Kimura, H. The possible role of hydrogen sulfide as an endogenous neuromodulator. J. Neurosci. 1996, 16, 1066–1071. [Google Scholar] [CrossRef] [Green Version]
- Guo, H.; Xiao, T.; Zhou, H.; Xie, Y.; Shen, W. Hydrogen sulfide: A versatile regulator of environmental stress in plants. Acta Physiol. Plant. 2016, 38, 16. [Google Scholar] [CrossRef]
- Aroca, A.; Gotor, C.; Bassham, D.C.; Romero, L.C. Hydrogen sulfide: From a toxic molecule to a key molecule of cell life. Antioxidants 2020, 9, 621. [Google Scholar] [CrossRef]
- Iciek, M.; Bilska-Wilkosz, A.; Górny, M. Sulfane sulfur-new findings on an old topic. Acta Biochim. Pol. 2019, 66, 533–544. [Google Scholar] [CrossRef] [PubMed]
- Bloem, E.; Haneklaus, S.; Kesselmeier, J.; Schnug, E. Sulfur fertilization and fungal infections affect the exchange of H2S and COS from agricultural crops. J. Agric. Food Chem. 2012, 60, 7588–7596. [Google Scholar] [CrossRef] [PubMed]
- Lisjak, M.; Teklic, T.; Wilson, I.D.; Whiteman, M.; Hancock, J.T. Hydrogen sulfide: Environmental factor or signalling molecule? Plant Cell Environ. 2013, 36, 1607–1616. [Google Scholar] [CrossRef] [PubMed]
- Gotor, C.; García, I.; Aroca, Á.; Laureano-Marín, A.M.; Arenas-Alfonseca, L.; Jurado-Flores, A.; Moreno, I.; Romero, L.C.; Kopriva, S. Signaling by hydrogen sulfide and cyanide through post-translational modification. J. Exp. Bot. 2019, 70, 4251–4265. [Google Scholar] [CrossRef]
- Aroca, Á.; Serna, A.; Gotor, C.; Romero, L.C. S-sulfhydration: A cysteine posttranslational modification in plant systems. Plant Physiol. 2015, 168, 334–342. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.G.; Xie, L.R.; Li, X.J. Hydrogen sulfide acts as a downstream signal molecule in salicylic acid-induced heat tolerance in maize (Zea mays L.) seedlings. J. Plant Physiol. 2015, 177, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Hou, Z.; Liu, J.; Hou, H.; Li, X.; Liu, X. H2S may function downstream of H2O2 in jasmonic acid-induced stomatal closure in Vicia faba. Chin. Bull. Bot. 2011, 46, 396–406. [Google Scholar] [CrossRef]
- Creelman, R.A.; Mullet, J.E. Biosynthesis and action of jasmonates in plants. Annu. Rev. Plant Biol. 1997, 48, 355–381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nazar, R.; Khan, M.I.R.; Iqbal, N.; Masood, A.; Khan, N.A. Involvement of ethylene in reversal of salt-inhibited photosynthesis by sulfur in mustard. Physiol. Plant. 2014, 152, 331–344. [Google Scholar] [CrossRef]
- Wawrzynska, A.; Moniuszko, G.; Sirko, A. Links between ethylene and sulfur nutrition—A regulatory interplay or just metabolite association? Front. Plant Sci. 2015, 6, 1053. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Hou, L.X.; Liu, G.H.; Liu, X.; Wang, X.C. Hydrogen sulfide induced by nitric oxide mediates ethylene-induced stomatal closure of Arabidopsis thaliana. Chin. Sci. Bull. 2011, 56, 3547–3553. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Yi, H. Differential expression of Arabidopsis defense-related genes in response to sulfur dioxide. Chemosphere 2012, 87, 718–724. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Yi, H. Genome-wide transcriptome analysis of Arabidopsis response to sulfur dioxide fumigation. Mol. Genet. Genom. 2014, 289, 989–999. [Google Scholar] [CrossRef] [PubMed]
- Brychkova, G.; Yarmolinsky, D.; Fluhr, R.; Sagi, M. The determination of sulfite levels and its oxidation in plant leaves. Plant Sci. 2012, 190, 123–130. [Google Scholar] [CrossRef]
- Yarmolinsky, D.; Brychkova, G.; Fluhr, R.; Sagi, M. Sulfite reductase protects plants against sulfite toxicity. Plant Physiol. 2013, 161, 725–743. [Google Scholar] [CrossRef] [Green Version]
- Brychkova, G.; Xia, Z.; Yang, G.; Yesbergenova, Z.; Zhang, Z.; Davydov, O.; Fluhr, R.; Sagi, M. Sulfite oxidase protects plants against sulfur dioxide toxicity. Plant J. 2007, 50, 696–709. [Google Scholar] [CrossRef]
- Xia, Z.; Su, X.; Wu, J.; Wu, K.; Zhang, H. Molecular cloning and functional characterization of a putative sulfite oxidase (SO) ortholog from Nicotiana benthamiana. Mol. Biol. Rep. 2012, 39, 2429–2437. [Google Scholar] [CrossRef]
- Zhang, X.; Wong, S.M. Hibiscus chlorotic ringspot virus upregulates plant sulfite oxidase transcripts and increases sulfate levels in kenaf (Hibiscus cannabinus L.). J. Gen. Virol. 2009, 90, 3042–3050. [Google Scholar] [CrossRef]
- Xue, M.; Yi, H. Enhanced Arabidopsis disease resistance against Botrytis cinerea induced by sulfur dioxide. Ecotoxicol. Environ. Saf. 2018, 147, 523–529. [Google Scholar] [CrossRef]
- Duarte-Sierra, A.; Aispuro-Hernández, E.; Vargas-Arispuro, I.; Islas-Osuna, M.A.; González-Aguilar, G.A.; Martínez-Téllez, M.Á. Quality and PR gene expression of table grapes treated with ozone and sulfur dioxide to control fungal decay. J. Sci. Food Agric. 2016, 96, 2018–2024. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Künstler, A.; Gullner, G.; Ádám, A.L.; Kolozsváriné Nagy, J.; Király, L. The Versatile Roles of Sulfur-Containing Biomolecules in Plant Defense—A Road to Disease Resistance. Plants 2020, 9, 1705. https://0-doi-org.brum.beds.ac.uk/10.3390/plants9121705
Künstler A, Gullner G, Ádám AL, Kolozsváriné Nagy J, Király L. The Versatile Roles of Sulfur-Containing Biomolecules in Plant Defense—A Road to Disease Resistance. Plants. 2020; 9(12):1705. https://0-doi-org.brum.beds.ac.uk/10.3390/plants9121705
Chicago/Turabian StyleKünstler, András, Gábor Gullner, Attila L. Ádám, Judit Kolozsváriné Nagy, and Lóránt Király. 2020. "The Versatile Roles of Sulfur-Containing Biomolecules in Plant Defense—A Road to Disease Resistance" Plants 9, no. 12: 1705. https://0-doi-org.brum.beds.ac.uk/10.3390/plants9121705