Role of Nickel in Microbial Pathogenesis
Abstract
:1. Introduction
2. Nickel Availability to Pathogens and Host-Mediated Influences
3. Ureases
3.1. H. pylori
3.2. S. aureus
3.3. P. mirabilis
3.4. Ureaplasma spp.
3.5. Eukaryotic Pathogens
4. Hydrogenases
4.1. H. pylori
4.2. H. hepaticus
4.3. S. Typhimurium
4.4. C. jejuni
4.5. C. concisus
4.6. S. flexneri
5. Other Ni-Dependent Enzymes
5.1. Acireductone Dioxygenase (ARD)
5.2. Ni-Glyoxalase I
5.3. Ni-Superoxide Dismutase (Ni-SOD)
6. Nickel Transport and Nickel Metallophores
7. Nickel Storage, Toxicity, and Metabolism
7.1. Hpn and Hpn-Like Proteins
7.2. HspA
8. Conclusions
Funding
Conflicts of Interest
References
- Zambelli, B.; Uversky, V.N.; Ciurli, S. Nickel impact on human health: An intrinsic disorder perspective. Biochim. Biophys. Acta (BBA) Proteins Proteom. 2016, 1864, 1714–1731. [Google Scholar] [CrossRef]
- Rutherford, J.C. The emerging role of urease as a general microbial virulence factor. PLoS Pathog. 2014, 10, e1004062. [Google Scholar] [CrossRef]
- Benoit, S.L.; Maier, R.J. Hydrogen and nickel metabolism in Helicobacter species. Ann. N. Y. Acad. Sci. 2008, 1125, 242–251. [Google Scholar] [CrossRef]
- Maier, R.J. Use of molecular hydrogen as an energy substrate by human pathogenic bacteria. Biochem. Soc. Trans. 2005, 33, 83–85. [Google Scholar] [CrossRef] [Green Version]
- Boer, J.L.; Mulrooney, S.B.; Hausinger, R.P. Nickel-dependent metalloenzymes. Arch. Biochem. Biophys. 2014, 544, 142–152. [Google Scholar] [CrossRef]
- Maroney, M.J.; Ciurli, S. Nonredox nickel enzymes. Chem. Rev. 2013, 114, 4206–4228. [Google Scholar] [CrossRef]
- Mazzei, L.; Musiani, F.; Ciurli, S. Urease. In The Biological Chemistry of Nickel; The Royal Society of Chemistry: Cambridge, UK, 2017; pp. 60–97. [Google Scholar]
- Tai, H.; Higuchi, Y.; Hirota, S. Comprehensive reaction mechanisms at and near the Ni–Fe active sites of [NiFe] hydrogenases. Dalton Trans. 2018, 47, 4408–4423. [Google Scholar] [CrossRef]
- Ogata, H.; Lubitz, W.; Higuchi, Y. Structure and function of [NiFe] hydrogenases. J. Biochem. 2016, 160, 251–258. [Google Scholar] [CrossRef] [Green Version]
- De Reuse, H.; Vinella, D.; Cavazza, C. Common themes and unique proteins for the uptake and trafficking of nickel, a metal essential for the virulence of Helicobacter pylori. Front. Cell. Infect. Microbiol. 2013, 3, 94. [Google Scholar] [CrossRef]
- Zeer-Wanklyn, C.J.; Zamble, D.B. Microbial nickel: Cellular uptake and delivery to enzyme centers. Curr. Opin. Chem. Biol. 2017, 37, 80–88. [Google Scholar] [CrossRef]
- Gaddy, J.A.; Haley, K.P. Metalloregulation of Helicobacter pylori physiology and pathogenesis. Front. Microbiol. 2015, 6, 911. [Google Scholar]
- Deshpande, A.R.; Pochapsky, T.C.; Ringe, D. The Metal Drives the Chemistry: Dual Functions of Acireductone Dioxygenase. Chem. Rev. 2017, 117, 10474–10501. [Google Scholar] [CrossRef]
- Miller, A.-F. Superoxide dismutases: Ancient enzymes and new insights. FEBS Lett. 2012, 586, 585–595. [Google Scholar] [CrossRef]
- Ryan, K.C.; Guce, A.I.; Johnson, O.E.; Brunold, T.C.; Cabelli, D.E.; Garman, S.C.; Maroney, M.J. Nickel superoxide dismutase: Structural and functional roles of His1 and its H-bonding network. Biochemistry 2015, 54, 1016–1027. [Google Scholar] [CrossRef]
- Honek, J.F. Nickel Glyoxalase I. In The Biological Chemistry of Nickel; Zamble, D., Rowinska-Zyrek, M., Kozlowski, H., Eds.; Royal Society of Chemistry: Cambridge, UK, 2017. [Google Scholar]
- Cox, G.M.; Mukherjee, J.; Cole, G.T.; Casadevall, A.; Perfect, J.R. Urease as a virulence factor in experimental cryptococcosis. Infect. Immun. 2000, 68, 443–448. [Google Scholar] [CrossRef]
- Olszewski, M.A.; Noverr, M.C.; Chen, G.-H.; Toews, G.B.; Cox, G.M.; Perfect, J.R.; Huffnagle, G.B. Urease expression by Cryptococcus neoformans promotes microvascular sequestration, thereby enhancing central nervous system invasion. Am. J. Pathol. 2004, 164, 1761–1771. [Google Scholar] [CrossRef]
- Fu, M.S.; Coelho, C.; De Leon-Rodriguez, C.M.; Rossi, D.C.P.; Camacho, E.; Jung, E.H.; Kulkarni, M.; Casadevall, A. Cryptococcus neoformans urease affects the outcome of intracellular pathogenesis by modulating phagolysosomal pH. PLoS Pathog. 2018, 14, e1007144. [Google Scholar] [CrossRef]
- Rodrigues, M.L.; Nakayasu, E.S.; Oliveira, D.L.; Nimrichter, L.; Nosanchuk, J.D.; Almeida, I.C.; Casadevall, A. Extracellular vesicles produced by Cryptococcus neoformans contain protein components associated with virulence. Eukaryot. Cell 2008, 7, 58–67. [Google Scholar] [CrossRef]
- Feder, V.; Kmetzsch, L.; Staats, C.C.; Vidal-Figueiredo, N.; Ligabue-Braun, R.; Carlini, C.R.; Vainstein, M.H. Cryptococcus gattii urease as a virulence factor and the relevance of enzymatic activity in cryptococcosis pathogenesis. FEBS J. 2015, 282, 1406–1418. [Google Scholar] [CrossRef]
- Mirbod-Donovan, F.; Schaller, R.; Hung, C.Y.; Xue, J.; Reichard, U.; Cole, G.T. Urease produced by Coccidioides posadasii contributes to the virulence of this respiratory pathogen. Infect. Immun. 2006, 74, 504–515. [Google Scholar] [CrossRef]
- Wise, H.Z.; Hung, C.-Y.; Whiston, E.; Taylor, J.W.; Cole, G.T. Extracellular ammonia at sites of pulmonary infection with Coccidioides posadasii contributes to severity of the respiratory disease. Microb. Pathog. 2013, 59, 19–28. [Google Scholar] [CrossRef]
- Baltazar, L.M.; Zamith-Miranda, D.; Burnet, M.C.; Choi, H.; Nimrichter, L.; Nakayasu, E.S.; Nosanchuk, J.D. Concentration-dependent protein loading of extracellular vesicles released by Histoplasma capsulatum after antibody treatment and its modulatory action upon macrophages. Sci. Rep. 2018, 8, 8065. [Google Scholar] [CrossRef]
- Costa, M.; Borges, C.L.; Bailao, A.M.; Meirelles, G.V.; Mendonça, Y.A.; Dantas, S.F.; de Faria, F.P.; Felipe, M.S.; Molinari-Madlum, E.N.E.; Mendes-Giannini, M.J. Transcriptome profiling of Paracoccidioides brasiliensis yeast-phase cells recovered from infected mice brings new insights into fungal response upon host interaction. Microbiology 2007, 153, 4194–4207. [Google Scholar] [CrossRef]
- Rujirawat, T.; Patumcharoenpol, P.; Lohnoo, T.; Yingyong, W.; Kumsang, Y.; Payattikul, P.; Tangphatsornruang, S.; Suriyaphol, P.; Reamtong, O.; Garg, G.; et al. Probing the Phylogenomics and Putative Pathogenicity Genes of Pythium insidiosum by Oomycete Genome Analyses. Sci. Rep. 2018, 8, 4135. [Google Scholar] [CrossRef]
- Ariza, A.; Vickers, T.J.; Greig, N.; Armour, K.A.; Dixon, M.J.; Eggleston, I.M.; Fairlamb, A.H.; Bond, C.S. Specificity of the trypanothione-dependent Leishmania major glyoxalase I: Structure and biochemical comparison with the human enzyme. Mol. Microbiol. 2006, 59, 1239–1248. [Google Scholar] [CrossRef]
- Chauhan, S.C.; Madhubala, R. Glyoxalase I gene deletion mutants of Leishmania donovani exhibit reduced methylglyoxal detoxification. PLoS ONE 2009, 4, e6805. [Google Scholar] [CrossRef]
- Greig, N.; Wyllie, S.; Vickers, T.J.; Fairlamb, A.H. Trypanothione-dependent glyoxalase I in Trypanosoma cruzi. Biochem. J. 2006, 400, 217–223. [Google Scholar] [CrossRef]
- Morou-Bermudez, E.; Burne, R.A. Genetic and physiologic characterization of urease of Actinomyces naeslundii. Infect. Immun. 1999, 67, 504–512. [Google Scholar]
- Salem, N.; Salem, L.; Saber, S.; Ismail, G.; Bluth, M.H. Corynebacterium urealyticum: A comprehensive review of an understated organism. Infect. Drug Resist. 2015, 8, 129–145. [Google Scholar]
- Sassetti, C.M.; Rubin, E.J. Genetic requirements for mycobacterial survival during infection. Proc. Natl. Acad. Sci. USA 2003, 100, 12989–12994. [Google Scholar] [CrossRef] [Green Version]
- Fontán, P.; Aris, V.; Ghanny, S.; Soteropoulos, P.; Smith, I. Global transcriptional profile of Mycobacterium tuberculosis during THP-1 human macrophage infection. Infect. Immun. 2008, 76, 717–725. [Google Scholar] [CrossRef]
- Schnappinger, D.; Ehrt, S.; Voskuil, M.I.; Liu, Y.; Mangan, J.A.; Monahan, I.M.; Dolganov, G.; Efron, B.; Butcher, P.D.; Nathan, C. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: Insights into the phagosomal environment. J. Exp. Med. 2003, 198, 693–704. [Google Scholar] [CrossRef]
- Lin, W.; Mathys, V.; Ang, E.L.; Koh, V.H.; Martinez Gomez, J.M.; Ang, M.L.; Zainul Rahim, S.Z.; Tan, M.P.; Pethe, K.; Alonso, S. Urease activity represents an alternative pathway for Mycobacterium tuberculosis nitrogen metabolism. Infect. Immun. 2012, 80, 2771–2779. [Google Scholar] [CrossRef]
- Suttisansanee, U.; Lau, K.; Lagishetty, S.; Rao, K.N.; Swaminathan, S.; Sauder, J.M.; Burley, S.K.; Honek, J.F. Structural Variation in Bacterial Glyoxalase I Enzymes Investigation of the Metalloenzyme Glyoxalase I from Clostridium acetobutylicum. J. Biol. Chem. 2011, 286, 38367–38374. [Google Scholar] [CrossRef]
- Resch, A.; Rosenstein, R.; Nerz, C.; Gotz, F. Differential gene expression profiling of Staphylococcus aureus cultivated under biofilm and planktonic conditions. Appl. Environ. Microbiol. 2005, 71, 2663–2676. [Google Scholar] [CrossRef]
- Zhou, C.; Bhinderwala, F.; Lehman, M.K.; Thomas, V.C.; Chaudhari, S.S.; Yamada, K.J.; Foster, K.W.; Powers, R.; Kielian, T.; Fey, P.D. Urease is an essential component of the acid response network of Staphylococcus aureus and is required for a persistent murine kidney infection. PLoS Pathog. 2019, 15, e1007538. [Google Scholar] [CrossRef]
- Vandecandelaere, I.; Van Nieuwerburgh, F.; Deforce, D.; Coenye, T. Metabolic activity, urease production, antibiotic resistance and virulence in dual species biofilms of Staphylococcus epidermidis and Staphylococcus aureus. PLoS ONE 2017, 12, e0172700. [Google Scholar] [CrossRef]
- Gatermann, S.; John, J.; Marre, R. Staphylococcus saprophyticus urease: Characterization and contribution to uropathogenicity in unobstructed urinary tract infection of rats. Infect. Immun. 1989, 57, 110–116. [Google Scholar]
- Chen, Y.Y.; Weaver, C.A.; Burne, R.A. Dual functions of Streptococcus salivarius urease. J. Bacteriol. 2000, 182, 4667–4669. [Google Scholar] [CrossRef]
- Kokkayil, P.; Dhawan, B. Ureaplasma: Current perspectives. Indian J. Med. Microbiol. 2015, 33, 205–214. [Google Scholar]
- Smith, D.; Russell, W.; Ingledew, W.; Thirkell, D. Hydrolysis of urea by Ureaplasma urealyticum generates a transmembrane potential with resultant ATP synthesis. J. Bacteriol. 1993, 175, 3253–3258. [Google Scholar] [CrossRef]
- Grenabo, L.; Hedelin, H.; Pettersson, S. Urinary infection stones caused by Ureaplasma urealyticum: A review. Scand. J. Infect. Dis. Suppl. 1988, 53, 46–49. [Google Scholar]
- Silva, J.; Marques, L.; Timenetsky, J.; de Farias, S.T. Ureaplasma diversum protein interaction networks: Evidence of horizontal gene transfer and evolution of reduced genomes among the Mollicutes. Can. J. Microbiol. 2019. [Google Scholar] [CrossRef]
- Sangari, F.J.; Seoane, A.; Rodriguez, M.C.; Aguero, J.; Garcia Lobo, J.M. Characterization of the urease operon of Brucella abortus and assessment of its role in virulence of the bacterium. Infect. Immun. 2007, 75, 774–780. [Google Scholar] [CrossRef]
- Abkar, M.; Amani, J.; Sahebghadam Lotfi, A.; Nikbakht Brujeni, G.; Alamian, S.; Kamali, M. Subcutaneous immunization with a novel immunogenic candidate (urease) confers protection against Brucella abortus and Brucella melitensis infections. APMIS 2015, 123, 667–675. [Google Scholar] [CrossRef]
- Bandara, A.B.; Contreras, A.; Contreras-Rodriguez, A.; Martins, A.M.; Dobrean, V.; Poff-Reichow, S.; Rajasekaran, P.; Sriranganathan, N.; Schurig, G.G.; Boyle, S.M. Brucella suis urease encoded by ure 1 but not ure 2 is necessary for intestinal infection of BALB/c mice. BMC Microbiol. 2007, 7, 57. [Google Scholar] [CrossRef]
- Smith, M.G.; Gianoulis, T.A.; Pukatzki, S.; Mekalanos, J.J.; Ornston, L.N.; Gerstein, M.; Snyder, M. New insights into Acinetobacter baumannii pathogenesis revealed by high-density pyrosequencing and transposon mutagenesis. Genes Dev. 2007, 21, 601–614. [Google Scholar] [CrossRef]
- Rathinavelu, S.; Zavros, Y.; Merchant, J.L. Acinetobacter lwoffii infection and gastritis. Microbes Infect. 2003, 5, 651–657. [Google Scholar] [CrossRef]
- Bossé, J.T.; MacInnes, J.I. Urease activity may contribute to the ability of Actinobacillus pleuropneumoniae to establish infection. Can. J. Vet. Res. 2000, 64, 145. [Google Scholar]
- Klitgaard, K.; Friis, C.; Jensen, T.K.; Angen, O.; Boye, M. Transcriptional portrait of Actinobacillus pleuropneumoniae during acute disease—Potential strategies for survival and persistence in the host. PLoS ONE 2012, 7, e35549. [Google Scholar] [CrossRef]
- Pinske, C.; Sawers, R.G. Anaerobic Formate and Hydrogen Metabolism. EcoSal Plus 2016, 7. [Google Scholar] [CrossRef]
- Steyert, S.R.; Kaper, J.B. Contribution of urease to colonization by Shiga toxin-producing Escherichia coli. Infect. Immun. 2012, 80, 2589–2600. [Google Scholar] [CrossRef]
- Li, M.F.; Sun, L. Edwardsiella tarda Sip2: A Serum-Induced Protein That Is Essential to Serum Survival, Acid Resistance, Intracellular Replication, and Host Infection. Front. Microbiol. 2018, 9, 1084. [Google Scholar] [CrossRef]
- Murphy, T.F.; Brauer, A.L. Expression of urease by Haemophilus influenzae during human respiratory tract infection and role in survival in an acid environment. BMC Microbiol. 2011, 11, 183. [Google Scholar] [CrossRef]
- Maroncle, N.; Rich, C.; Forestier, C. The role of Klebsiella pneumoniae urease in intestinal colonization and resistance to gastrointestinal stress. Res. Microbiol. 2006, 157, 184–193. [Google Scholar] [CrossRef]
- Young, G.M.; Amid, D.; Miller, V.L. A bifunctional urease enhances survival of pathogenic Yersinia enterocolitica and Morganella morganii at low pH. J. Bacteriol. 1996, 178, 6487–6495. [Google Scholar] [CrossRef]
- Armbruster, C.E.; Smith, S.N.; Yep, A.; Mobley, H.L. Increased incidence of urolithiasis and bacteremia during Proteus mirabilis and Providencia stuartii coinfection due to synergistic induction of urease activity. J. Infect. Dis. 2014, 209, 1524–1532. [Google Scholar] [CrossRef]
- Alteri, C.J.; Himpsl, S.D.; Engstrom, M.D.; Mobley, H.L. Anaerobic respiration using a complete oxidative TCA cycle drives multicellular swarming in Proteus mirabilis. MBio 2012, 3, e00365-12. [Google Scholar] [CrossRef]
- Johnson, D.; Russell, R.; Lockatell, C.; Zulty, J.; Warren, J.; Mobley, H. Contribution of Proteus mirabilis urease to persistence, urolithiasis, and acute pyelonephritis in a mouse model of ascending urinary tract infection. Infect. Immun. 1993, 61, 2748–2754. [Google Scholar]
- Schaffer, J.N.; Norsworthy, A.N.; Sun, T.T.; Pearson, M.M. Proteus mirabilis fimbriae- and urease-dependent clusters assemble in an extracellular niche to initiate bladder stone formation. Proc. Natl. Acad. Sci. USA 2016, 113, 4494–4499. [Google Scholar] [CrossRef]
- Armbruster, C.E.; Mobley, H.L.T.; Pearson, M.M. Pathogenesis of Proteus mirabilis Infection. EcoSal Plus 2018, 8. [Google Scholar] [CrossRef] [PubMed]
- Zbell, A.L.; Maier, S.E.; Maier, R.J. Salmonella enterica serovar Typhimurium NiFe uptake-type hydrogenases are differentially expressed in vivo. Infect. Immun. 2008, 76, 4445–4454. [Google Scholar] [CrossRef] [PubMed]
- Maier, L.; Barthel, M.; Stecher, B.; Maier, R.J.; Gunn, J.S.; Hardt, W.-D. Salmonella Typhimurium strain ATCC14028 requires H2-hydrogenases for growth in the gut, but not at systemic sites. PLoS ONE 2014, 9, e110187. [Google Scholar] [CrossRef] [PubMed]
- Lam, L.H.; Monack, D.M. Intraspecies competition for niches in the distal gut dictate transmission during persistent Salmonella infection. PLoS Pathog. 2014, 10, e1004527. [Google Scholar] [CrossRef] [PubMed]
- Zbell, A.L.; Benoit, S.L.; Maier, R.J. Differential expression of NiFe uptake-type hydrogenase genes in Salmonella enterica serovar Typhimurium. Microbiology 2007, 153, 3508–3516. [Google Scholar] [CrossRef] [PubMed]
- Parkin, A.; Bowman, L.; Roessler, M.M.; Davies, R.A.; Palmer, T.; Armstrong, F.A.; Sargent, F. How Salmonella oxidises H2 under aerobic conditions. FEBS Lett. 2012, 586, 536–544. [Google Scholar] [CrossRef] [PubMed]
- Lamichhane-Khadka, R.; Benoit, S.L.; Miller-Parks, E.F.; Maier, R.J. Host hydrogen rather than that produced by the pathogen is important for Salmonella enterica serovar Typhimurium virulence. Infect. Immun. 2015, 83, 311–316. [Google Scholar] [CrossRef]
- McNorton, M.M.; Maier, R.J. Roles of H2 uptake hydrogenases in Shigella flexneri acid tolerance. Microbiology 2012, 158, 2204–2212. [Google Scholar] [CrossRef]
- Cai, Y.; Ni, Y. Purification, characterization, and pathogenicity of urease produced by Vibrio parahaemolyticus. J. Clin. Lab. Anal. 1996, 10, 70–73. [Google Scholar] [CrossRef]
- De Koning-Ward, T.F.; Robins-Browne, R.M. Contribution of urease to acid tolerance in Yersinia enterocolitica. Infect. Immun. 1995, 63, 3790–3795. [Google Scholar]
- Da Silva, S.M.; Venceslau, S.S.; Fernandes, C.L.; Valente, F.M.; Pereira, I.A. Hydrogen as an energy source for the human pathogen Bilophila wadsworthia. Antonie Leeuwenhoek 2008, 93, 381–390. [Google Scholar] [CrossRef] [PubMed]
- Weerakoon, D.R.; Borden, N.J.; Goodson, C.M.; Grimes, J.; Olson, J.W. The role of respiratory donor enzymes in Campylobacter jejuni host colonization and physiology. Microb. Pathog. 2009, 47, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Kassem, I.; Khatri, M.; Esseili, M.A.; Sanad, Y.M.; Saif, Y.M.; Olson, J.W.; Rajashekara, G. Respiratory proteins contribute differentially to Campylobacter jejuni’s survival and in vitro interaction with hosts’ intestinal cells. BMC Microbiol. 2012, 12, 258. [Google Scholar] [CrossRef] [PubMed]
- Benoit, S.L.; Maier, R.J. Site-directed mutagenesis of Campylobacter concisus respiratory genes provides insight into the pathogen’s growth requirements. Sci. Rep. 2018, 8, 14203. [Google Scholar] [CrossRef] [PubMed]
- Mehta, N.S.; Benoit, S.; Mysore, J.V.; Sousa, R.S.; Maier, R.J. Helicobacter hepaticus hydrogenase mutants are deficient in hydrogen-supported amino acid uptake and in causing liver lesions in A/J mice. Infect. Immun. 2005, 73, 5311–5318. [Google Scholar] [CrossRef] [PubMed]
- Ge, Z.; Lee, A.; Whary, M.T.; Rogers, A.B.; Maurer, K.J.; Taylor, N.S.; Schauer, D.B.; Fox, J.G. Helicobacter hepaticus urease is not required for intestinal colonization but promotes hepatic inflammation in male A/JCr mice. Microb. Pathog. 2008, 45, 18–24. [Google Scholar] [CrossRef]
- Andrutis, K.A.; Fox, J.G.; Schauer, D.B.; Marini, R.P.; Murphy, J.C.; Yan, L.; Solnick, J.V. Inability of an isogenic urease-negative mutant stain of Helicobacter mustelae to colonize the ferret stomach. Infect. Immun. 1995, 63, 3722–3725. [Google Scholar]
- Olson, J.W.; Maier, R.J. Molecular hydrogen as an energy source for Helicobacter pylori. Science 2002, 298, 1788–1790. [Google Scholar] [CrossRef]
- Kuhns, L.G.; Benoit, S.L.; Bayyareddy, K.; Johnson, D.; Orlando, R.; Evans, A.L.; Waldrop, G.L.; Maier, R.J. Carbon fixation driven by Molecular Hydrogen Results in Chemolithoautotrophically Enhanced Growth of Helicobacter pylori. J. Bacteriol. 2016, 198, 1423–1428. [Google Scholar] [CrossRef]
- Wang, G.; Romero-Gallo, J.; Benoit, S.L.; Piazuelo, M.B.; Dominguez, R.L.; Morgan, D.R.; Peek, R.M., Jr.; Maier, R.J. Hydrogen metabolism in Helicobacter pylori plays a role in gastric carcinogenesis through facilitating CagA translocation. MBio 2016, 7, e01022-16. [Google Scholar] [CrossRef]
- Segal, E.D.; Shon, J.; Tompkins, L.S. Characterization of Helicobacter pylori urease mutants. Infect. Immun. 1992, 60, 1883–1889. [Google Scholar] [PubMed]
- Tsuda, M.; Karita, M.; Morshed, M.G.; Okita, K.; Nakazawa, T. A urease-negative mutant of Helicobacter pylori constructed by allelic exchange mutagenesis lacks the ability to colonize the nude mouse stomach. Infect. Immun. 1994, 62, 3586–3589. [Google Scholar] [PubMed]
- Eaton, K.A.; Krakowka, S. Effect of gastric pH on urease-dependent colonization of gnotobiotic piglets by Helicobacter pylori. Infect. Immun. 1994, 62, 3604–3607. [Google Scholar] [PubMed]
- Harris, P.R.; Ernst, P.B.; Kawabata, S.; Kiyono, H.; Graham, M.F.; Smith, P.D. Recombinant Helicobacter pylori urease activates primary mucosal macrophages. J. Infect. Dis. 1998, 178, 1516–1520. [Google Scholar] [CrossRef] [PubMed]
- Harris, P.; Mobley, H.; Perez-Perez, G.; Blaser, M.; Smith, P. Helicobacter pylori urease is a potent stimulus of mononuclear phagocyte activation and inflammatory cytokine production. Gastroenterology 1996, 111, 419–425. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Gunasena, H.; Cheng, Z.; Espejo, R.; Crowe, S.E.; Ernst, P.B.; Reyes, V.E. Helicobacter pylori urease binds to class II MHC on gastric epithelial cells and induces their apoptosis. J. Immunol. 2000, 165, 1918–1924. [Google Scholar] [CrossRef] [PubMed]
- Kavermann, H.; Burns, B.P.; Angermuller, K.; Odenbreit, S.; Fischer, W.; Melchers, K.; Haas, R. Identification and characterization of Helicobacter pylori genes essential for gastric colonization. J. Exp. Med. 2003, 197, 813–822. [Google Scholar] [CrossRef]
- Wirth, H.P.; Beins, M.H.; Yang, M.; Tham, K.T.; Blaser, M.J. Experimental infection of Mongolian gerbils with wild-type and mutant Helicobacter pylori strains. Infect. Immun. 1998, 66, 4856–4866. [Google Scholar]
- Kuwahara, H.; Miyamoto, Y.; Akaike, T.; Kubota, T.; Sawa, T.; Okamoto, S.; Maeda, H. Helicobacter pylori urease suppresses bactericidal activity of peroxynitrite via carbon dioxide production. Infect. Immun. 2000, 68, 4378–4383. [Google Scholar] [CrossRef]
- Lytton, S.D.; Fischer, W.; Nagel, W.; Haas, R.; Beck, F.X. Production of ammonium by Helicobacter pylori mediates occludin processing and disruption of tight junctions in Caco-2 cells. Microbiology 2005, 151, 3267–3276. [Google Scholar] [CrossRef]
- Wroblewski, L.E.; Shen, L.; Ogden, S.; Romero-Gallo, J.; Lapierre, L.A.; Israel, D.A.; Turner, J.R.; Peek, R.M., Jr. Helicobacter pylori dysregulation of gastric epithelial tight junctions by urease-mediated myosin II activation. Gastroenterology 2009, 136, 236–246. [Google Scholar] [CrossRef] [PubMed]
- Wassermann, G.E.; Olivera-Severo, D.; Uberti, A.F.; Carlini, C.R. Helicobacter pylori urease activates blood platelets through a lipoxygenase-mediated pathway. J. Cell. Mol. Med. 2010, 14, 2025–2034. [Google Scholar] [CrossRef] [PubMed]
- Perrais, M.; Rousseaux, C.; Ducourouble, M.-P.; Courcol, R.; Vincent, P.; Jonckheere, N.; Van Seuningen, I. Helicobacter pylori urease and flagellin alter mucin gene expression in human gastric cancer cells. Gastric Cancer 2014, 17, 235–246. [Google Scholar] [CrossRef] [PubMed]
- Debowski, A.W.; Walton, S.M.; Chua, E.G.; Tay, A.C.; Liao, T.; Lamichhane, B.; Himbeck, R.; Stubbs, K.A.; Marshall, B.J.; Fulurija, A.; et al. Helicobacter pylori gene silencing in vivo demonstrates urease is essential for chronic infection. PLoS Pathog. 2017, 13, e1006464. [Google Scholar] [CrossRef] [PubMed]
- Olivera-Severo, D.; Uberti, A.F.; Marques, M.S.; Pinto, M.T.; Gomez-Lazaro, M.; Figueiredo, C.; Leite, M.; Carlini, C.R. A new role for Helicobacter pylori urease: Contributions to angiogenesis. Front. Microbiol. 2017, 8, 1883. [Google Scholar] [CrossRef] [PubMed]
- Scopel-Guerra, A.; Olivera-Severo, D.; Staniscuaski, F.; Uberti, A.F.; Callai-Silva, N.; Jaeger, N.; Porto, B.N.; Carlini, C.R. The impact of Helicobacter pylori urease upon platelets and consequent contributions to inflammation. Front. Microbiol. 2017, 8, 2447. [Google Scholar] [CrossRef] [PubMed]
- Schmalstig, A.A.; Benoit, S.L.; Misra, S.K.; Sharp, J.S.; Maier, R.J. Noncatalytic antioxidant role for Helicobacter pylori urease. J. Bacteriol. 2018, 200, e00124-18. [Google Scholar] [CrossRef] [PubMed]
- Iyengar, G.V.; Kollmer, W.E.; Bowen, H.J.M. The Elemental Composition of Human Tissues and Body Fluids: A Compilation of Values for Adults; Verlag Chemie: Weinheim, Germany, 1978. [Google Scholar]
- Maret, W. Metalloproteomics, metalloproteomes, and the annotation of metalloproteins. Metallomics 2010, 2, 117–125. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Ochoa, V.E.; Jellbauer, S.; Klaus, S.; Raffatellu, M. Transition metal ions at the crossroads of mucosal immunity and microbial pathogenesis. Front. Cell. Infect. Microbiol. 2014, 4, 2. [Google Scholar] [CrossRef] [Green Version]
- Zackular, J.P.; Chazin, W.J.; Skaar, E.P. Nutritional Immunity: S100 Proteins at the host-pathogen interface. J. Biol. Chem. 2015, 290, 18991–18998. [Google Scholar] [CrossRef]
- Choby, J.E.; Mike, L.A.; Mashruwala, A.A.; Dutter, B.F.; Dunman, P.M.; Sulikowski, G.A.; Boyd, J.M.; Skaar, E.P. A small-molecule inhibitor of iron-sulfur cluster assembly uncovers a link between virulence regulation and metabolism in Staphylococcus aureus. Cell Chem. Biol. 2016, 23, 1351–1361. [Google Scholar] [CrossRef] [PubMed]
- Palmer, L.D.; Skaar, E.P. Transition Metals and Virulence in Bacteria. Annu. Rev. Genet. 2016, 50, 67–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakashige, T.G.; Zygiel, E.M.; Drennan, C.L.; Nolan, E.M. Nickel sequestration by the host-defense protein human calprotectin. J. Am. Chem. Soc. 2017, 139, 8828–8836. [Google Scholar] [CrossRef] [PubMed]
- Kanwar, J.R.; Roy, K.; Patel, Y.; Zhou, S.F.; Singh, M.R.; Singh, D.; Nasir, M.; Sehgal, R.; Sehgal, A.; Singh, R.S.; et al. Multifunctional iron bound lactoferrin and nanomedicinal approaches to enhance its bioactive functions. Molecules 2015, 20, 9703–9731. [Google Scholar] [CrossRef] [PubMed]
- Ganz, T. Iron and infection. Int. J. Hematol. 2018, 107, 7–15. [Google Scholar] [CrossRef]
- Kulprachakarn, K.; Chen, Y.L.; Kong, X.; Arno, M.C.; Hider, R.C.; Srichairatanakool, S.; Bansal, S.S. Copper(II) binding properties of hepcidin. J. Biol. Inorg. Chem. 2016, 21, 329–338. [Google Scholar] [CrossRef] [Green Version]
- Stefanova, D.; Raychev, A.; Deville, J.; Humphries, R.; Campeau, S.; Ruchala, P.; Nemeth, E.; Ganz, T.; Bulut, Y. Hepcidin protects against lethal Escherichia coli sepsis in mice inoculated with isolates from septic patients. Infect. Immun. 2018, 86, e00253-18. [Google Scholar] [CrossRef]
- Stefanova, D.; Raychev, A.; Arezes, J.; Ruchala, P.; Gabayan, V.; Skurnik, M.; Dillon, B.J.; Horwitz, M.A.; Ganz, T.; Bulut, Y.; et al. Endogenous hepcidin and its agonist mediate resistance to selected infections by clearing non-transferrin-bound iron. Blood 2017, 130, 245–257. [Google Scholar] [CrossRef]
- Stafford, S.L.; Bokil, N.J.; Achard, M.E.; Kapetanovic, R.; Schembri, M.A.; McEwan, A.G.; Sweet, M.J. Metal ions in macrophage antimicrobial pathways: Emerging roles for zinc and copper. Biosci. Rep. 2013, 33, e00049. [Google Scholar] [CrossRef]
- Cellier, M.F.M. Developmental Control of NRAMP1 (SLC11A1) Expression in Professional Phagocytes. Biology 2017, 6, 28. [Google Scholar] [CrossRef]
- Corbin, B.D.; Seeley, E.H.; Raab, A.; Feldmann, J.; Miller, M.R.; Torres, V.J.; Anderson, K.L.; Dattilo, B.M.; Dunman, P.M.; Gerads, R.; et al. Metal chelation and inhibition of bacterial growth in tissue abscesses. Science 2008, 319, 962–965. [Google Scholar] [CrossRef] [PubMed]
- Rowinska-Zyrek, M.; Zakrzewska-Czerwinska, J.; Zawilak-Pawlik, A.; Kozlowski, H. Ni2+ chemistry in pathogens–a possible target for eradication. Dalton Trans. 2014, 43, 8976–8989. [Google Scholar] [CrossRef] [PubMed]
- Maier, R.J. Availability and use of molecular hydrogen as an energy substrate for Helicobacter species. Microbes Infect. 2003, 5, 1159–1163. [Google Scholar] [CrossRef] [PubMed]
- Morrow, C.A.; Fraser, J.A. Is the nickel-dependent urease complex of Cryptococcus the pathogen’s Achilles’ heel? MBio 2013, 4, e00408-13. [Google Scholar] [CrossRef] [PubMed]
- Mora, D.; Arioli, S. Microbial urease in health and disease. PLoS Pathog. 2014, 10, e1004472. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.-T.; Chen, M.-L.; Huang, L.-L.; Mao, I.-F. Uric acid and urea in human sweat. Chin. J. Physiol. 2002, 45, 109–116. [Google Scholar] [PubMed]
- Lasisi, T.J.; Raji, Y.R.; Salako, B.L. Salivary creatinine and urea analysis in patients with chronic kidney disease: A case control study. BMC Nephrol. 2016, 17, 10. [Google Scholar] [CrossRef] [PubMed]
- Bishai, W.; Timmins, G. Potential for breath test diagnosis of urease positive pathogens in lung infections. J. Breath Res. 2019, 13, 032002. [Google Scholar] [CrossRef] [PubMed]
- Anderson, K.L.; Roux, C.M.; Olson, M.W.; Luong, T.T.; Lee, C.Y.; Olson, R.; Dunman, P.M. Characterizing the effects of inorganic acid and alkaline shock on the Staphylococcus aureus transcriptome and messenger RNA turnover. FEMS Immunol. Med. Microbiol. 2010, 60, 208–250. [Google Scholar] [CrossRef] [PubMed]
- Pot, R.G.; Stoof, J.; Nuijten, P.J.; De Haan, L.A.; Loeffen, P.; Kuipers, E.J.; Van Vliet, A.H.; Kusters, J.G. UreA2B2: A second urease system in the gastric pathogen Helicobacter felis. FEMS Immunol. Med. Microbiol. 2007, 50, 273–279. [Google Scholar] [CrossRef] [PubMed]
- Stoof, J.; Breijer, S.; Pot, R.G.; van der Neut, D.; Kuipers, E.J.; Kusters, J.G.; van Vliet, A.H. Inverse nickel-responsive regulation of two urease enzymes in the gastric pathogen Helicobacter mustelae. Environ. Microbiol. 2008, 10, 2586–2597. [Google Scholar] [CrossRef] [PubMed]
- Carter, E.L.; Tronrud, D.E.; Taber, S.R.; Karplus, P.A.; Hausinger, R.P. Iron-containing urease in a pathogenic bacterium. Proc. Natl. Acad. Sci. USA 2011, 108, 13095–13099. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Solomons, N.W.; Viteri, F.; Shuler, T.R.; Nielsen, F.H. Bioavailability of nickel in man: Effects of foods and chemically-defined dietary constituents on the absorption of inorganic nickel. J. Nutr. 1982, 112, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Bauerfeind, P.; Garner, R.; Dunn, B.; Mobley, H. Synthesis and activity of Helicobacter pylori urease and catalase at low pH. Gut 1997, 40, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, A.; Gupta, A.; Chandra, M.; Koowar, S. Role of Helicobacter pylori infection in the pathogenesis of minimal hepatic encephalopathy and effect of its eradication. Indian J. Gastroenterol. 2011, 30, 29–32. [Google Scholar] [CrossRef] [PubMed]
- Stingl, K.; De Reuse, H. Staying alive overdosed: How does Helicobacter pylori control urease activity? Int. J. Med. Microbiol. 2005, 295, 307–315. [Google Scholar] [CrossRef] [PubMed]
- van Vliet, A.H.; Poppelaars, S.W.; Davies, B.J.; Stoof, J.; Bereswill, S.; Kist, M.; Penn, C.W.; Kuipers, E.J.; Kusters, J.G. NikR mediates nickel-responsive transcriptional induction of urease expression in Helicobacter pylori. Infect. Immun. 2002, 70, 2846–2852. [Google Scholar] [CrossRef]
- van Vliet, A.H.; Kuipers, E.J.; Waidner, B.; Davies, B.J.; de Vries, N.; Penn, C.W.; Vandenbroucke-Grauls, C.M.; Kist, M.; Bereswill, S.; Kusters, J.G. Nickel-responsive induction of urease expression in Helicobacter pylori is mediated at the transcriptional level. Infect. Immun. 2001, 69, 4891–4897. [Google Scholar] [CrossRef] [PubMed]
- Tong, S.Y.; Davis, J.S.; Eichenberger, E.; Holland, T.L.; Fowler, V.G. Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef] [PubMed]
- Molina-Manso, D.; del Prado, G.; Ortiz-Perez, A.; Manrubia-Cobo, M.; Gomez-Barrena, E.; Cordero-Ampuero, J.; Esteban, J. In vitro susceptibility to antibiotics of staphylococci in biofilms isolated from orthopaedic infections. Int. J. Antimicrob. Agents 2013, 41, 521–523. [Google Scholar] [CrossRef]
- Norsworthy, A.N.; Pearson, M.M. From Catheter to Kidney Stone: The uropathogenic lifestyle of Proteus mirabilis. Trends Microbiol. 2017, 25, 304–315. [Google Scholar] [CrossRef] [PubMed]
- Bichler, K.H.; Eipper, E.; Naber, K.; Braun, V.; Zimmermann, R.; Lahme, S. Urinary infection stones. Int. J. Antimicrob. Agents 2002, 19, 488–498. [Google Scholar] [CrossRef]
- Heitland, P.; Köster, H.D. Biomonitoring of 30 trace elements in urine of children and adults by ICP-MS. Clin. Chim. Acta 2006, 365, 310–318. [Google Scholar] [CrossRef] [PubMed]
- Olson, J.W.; Mehta, N.S.; Maier, R.J. Requirement of nickel metabolism proteins HypA and HypB for full activity of both hydrogenase and urease in Helicobacter pylori. Mol. Microbiol. 2001, 39, 176–182. [Google Scholar] [CrossRef] [PubMed]
- Benoit, S.L.; Zbell, A.L.; Maier, R.J. Nickel enzyme maturation in Helicobacter hepaticus: Roles of accessory proteins in hydrogenase and urease activities. Microbiology 2007, 153, 3748–3756. [Google Scholar] [CrossRef] [PubMed]
- Pearson, M.M.; Sebaihia, M.; Churcher, C.; Quail, M.A.; Seshasayee, A.S.; Luscombe, N.M.; Abdellah, Z.; Arrosmith, C.; Atkin, B.; Chillingworth, T.; et al. Complete genome sequence of uropathogenic Proteus mirabilis, a master of both adherence and motility. J. Bacteriol. 2008, 190, 4027–4037. [Google Scholar] [CrossRef] [PubMed]
- Gerwien, F.; Skrahina, V.; Kasper, L.; Hube, B.; Brunke, S. Metals in fungal virulence. FEMS Microbiol. Rev. 2018, 42. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Panting, R.J.; Varma, A.; Saijo, T.; Waldron, K.J.; Jong, A.; Ngamskulrungroj, P.; Chang, Y.C.; Rutherford, J.C.; Kwon-Chung, K.J. Factors required for activation of urease as a virulence determinant in Cryptococcus neoformans. MBio 2013, 4, e00220-13. [Google Scholar] [CrossRef]
- Lubitz, W.; Ogata, H.; Rüdiger, O.; Reijerse, E. Hydrogenases. Chem. Rev. 2014, 114, 4081–4148. [Google Scholar] [CrossRef]
- Maier, R.J.; Olczak, A.; Maier, S.; Soni, S.; Gunn, J. Respiratory hydrogen use by Salmonella enterica serovar Typhimurium is essential for virulence. Infect. Immun. 2004, 72, 6294–6299. [Google Scholar] [CrossRef]
- Maier, R.J.; Fu, C.; Gilbert, J.; Moshiri, F.; Olson, J.; Plaut, A.G. Hydrogen uptake hydrogenase in Helicobacter pylori. FEMS Microbiol. Lett. 1996, 141, 71–76. [Google Scholar] [CrossRef] [PubMed]
- Ernst, F.D.; Bereswill, S.; Waidner, B.; Stoof, J.; Mader, U.; Kusters, J.G.; Kuipers, E.J.; Kist, M.; van Vliet, A.H.; Homuth, G. Transcriptional profiling of Helicobacter pylori Fur- and iron-regulated gene expression. Microbiology 2005, 151, 533–546. [Google Scholar] [CrossRef] [PubMed]
- Muller, C.; Bahlawane, C.; Aubert, S.; Delay, C.M.; Schauer, K.; Michaud-Soret, I.; De Reuse, H. Hierarchical regulation of the NikR-mediated nickel response in Helicobacter pylori. Nucleic Acids Res. 2011, 39, 7564–7575. [Google Scholar] [CrossRef] [PubMed]
- Contreras, M.; Thiberge, J.M.; Mandrand-Berthelot, M.A.; Labigne, A. Characterization of the roles of NikR, a nickel-responsive pleiotropic autoregulator of Helicobacter pylori. Mol. Microbiol. 2003, 49, 947–963. [Google Scholar] [CrossRef] [PubMed]
- Benoit, S.L.; Mehta, N.; Weinberg, M.V.; Maier, C.; Maier, R.J. Interaction between the Helicobacter pylori accessory proteins HypA and UreE is needed for urease maturation. Microbiology 2007, 153, 1474–1482. [Google Scholar] [CrossRef] [PubMed]
- Benoit, S.L.; McMurry, J.L.; Hill, S.A.; Maier, R.J. Helicobacter pylori hydrogenase accessory protein HypA and urease accessory protein UreG compete with each other for UreE recognition. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2012, 1820, 1519–1525. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.Q.; Huang, H.-T.; Maroney, M.J. The Helicobacter pylori HypA· UreE2 Complex Contains a Novel High-Affinity Ni(II)-Binding Site. Biochemistry 2018, 57, 2932–2942. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Li, H.; Cheng, T.; Xia, W.; Lai, Y.-T.; Sun, H. Nickel translocation between metallochaperones HypA and UreE in Helicobacter pylori. Metallomics 2014, 6, 1731–1736. [Google Scholar] [CrossRef]
- Stingl, K.; Schauer, K.; Ecobichon, C.; Labigne, A.; Lenormand, P.; Rousselle, J.-C.; Namane, A.; de Reuse, H. In vivo interactome of Helicobacter pylori urease revealed by tandem affinity purification. Mol. Cell. Proteom. 2008, 7, 2429–2441. [Google Scholar] [CrossRef]
- Levitt, M.D. Production and excretion of hydrogen gas in man. N. Engl. J. Med. 1969, 281, 122–127. [Google Scholar] [CrossRef]
- Kanazuru, T.; Sato, E.F.; Nagata, K.; Matsui, H.; Watanabe, K.; Kasahara, E.; Jikumaru, M.; Inoue, J.; Inoue, M. Role of hydrogen generation by Klebsiella pneumoniae in the oral cavity. J. Microbiol. 2010, 48, 778–783. [Google Scholar] [CrossRef] [PubMed]
- Blaser, M.J.; Perez-Perez, G.I.; Kleanthous, H.; Cover, T.L.; Peek, R.M.; Chyou, P.H.; Stemmermann, G.N.; Nomura, A. Infection with Helicobacter pylori strains possessing cagA is associated with an increased risk of developing adenocarcinoma of the stomach. Cancer Res. 1995, 55, 2111–2115. [Google Scholar] [PubMed]
- Fox, J.G.; Ge, Z.; Whary, M.T.; Erdman, S.E.; Horwitz, B.H. Helicobacter hepaticus infection in mice: Models for understanding lower bowel inflammation and cancer. Mucosal Immunol. 2011, 4, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Poutahidis, T.; Cappelle, K.; Levkovich, T.; Lee, C.W.; Doulberis, M.; Ge, Z.; Fox, J.G.; Horwitz, B.H.; Erdman, S.E. Pathogenic intestinal bacteria enhance prostate cancer development via systemic activation of immune cells in mice. PLoS ONE 2013, 8, e73933. [Google Scholar] [CrossRef] [PubMed]
- Suerbaum, S.; Josenhans, C.; Sterzenbach, T.; Drescher, B.; Brandt, P.; Bell, M.; Droge, M.; Fartmann, B.; Fischer, H.P.; Ge, Z.; et al. The complete genome sequence of the carcinogenic bacterium Helicobacter hepaticus. Proc. Natl. Acad. Sci. USA 2003, 100, 7901–7906. [Google Scholar] [CrossRef] [PubMed]
- Maier, R.J.; Olson, J.; Olczak, A. Hydrogen-oxidizing capabilities of Helicobacter hepaticus and in vivo availability of the substrate. J. Bacteriol. 2003, 185, 2680–2682. [Google Scholar] [CrossRef] [PubMed]
- McDowall, J.S.; Murphy, B.J.; Haumann, M.; Palmer, T.; Armstrong, F.A.; Sargent, F. Bacterial formate hydrogenlyase complex. Proc. Natl. Acad. Sci. USA 2014, 111, E3948–E3956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zbell, A.L.; Maier, R.J. Role of the Hya hydrogenase in recycling of anaerobically produced H2 in Salmonella enterica serovar Typhimurium. Appl. Environ. Microbiol. 2009, 75, 1456–1459. [Google Scholar] [CrossRef]
- Maier, L.; Vyas, R.; Cordova, C.D.; Lindsay, H.; Schmidt, T.S.; Brugiroux, S.; Periaswamy, B.; Bauer, R.; Sturm, A.; Schreiber, F.; et al. Microbiota-derived hydrogen fuels Salmonella typhimurium invasion of the gut ecosystem. Cell Host Microbe 2013, 14, 641–651. [Google Scholar] [CrossRef]
- Craig, M.; Sadik, A.Y.; Golubeva, Y.A.; Tidhar, A.; Slauch, J.M. Twin-arginine translocation system (tat) mutants of Salmonella are attenuated due to envelope defects, not respiratory defects. Mol. Microbiol. 2013, 89, 887–902. [Google Scholar] [CrossRef]
- Parkhill, J.; Wren, B.W.; Mungall, K.; Ketley, J.M.; Churcher, C.; Basham, D.; Chillingworth, T.; Davies, R.M.; Feltwell, T.; Holroyd, S.; et al. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 2000, 403, 665–668. [Google Scholar] [CrossRef]
- Carlone, G.M.; Lascelles, J. Aerobic and anaerobic respiratory systems in Campylobacter fetus subsp. jejuni grown in atmospheres containing hydrogen. J. Bacteriol. 1982, 152, 306–314. [Google Scholar]
- Istivan, T.; Ward, P.; Coloe, P. Current Research, Technology and Education Topics in Applied Microbiology and Microbial Biotechnology; Méndez-Vilas, A., Ed.; FORMATEX: Badajoz, Spain, 2010; pp. 626–634. [Google Scholar]
- Benoit, S.L.; Holland, A.A.; Johnson, M.K.; Maier, R.J. Iron-sulfur protein maturation in Helicobacter pylori: Identifying a Nfu-type cluster carrier protein and its iron-sulfur protein targets. Mol. Microbiol. 2018, 108, 379–396. [Google Scholar] [CrossRef]
- Lee, H.; Ma, R.; Grimm, M.C.; Riordan, S.M.; Lan, R.; Zhong, L.; Raftery, M.; Zhang, L. Examination of the anaerobic growth of Campylobacter concisus strains. Int. J. Microbiol. 2014, 2014, 476047. [Google Scholar] [CrossRef]
- Kotloff, K.L.; Riddle, M.S.; Platts-Mills, J.A.; Pavlinac, P.; Zaidi, A.K. Shigellosis. Lancet 2018, 391, 801–812. [Google Scholar] [CrossRef]
- Kissing, S.; Saftig, P.; Haas, A. Vacuolar ATPase in phago (lyso) some biology. Int. J. Med. Microbiol. 2018, 308, 58–67. [Google Scholar] [CrossRef]
- Pochapsky, T.C.; Pochapsky, S.S.; Ju, T.; Mo, H.; Al-Mjeni, F.; Maroney, M.J. Modeling and experiment yields the structure of acireductone dioxygenase from Klebsiella pneumoniae. Nat. Struct. Mol. Biol. 2002, 9, 966. [Google Scholar] [CrossRef]
- Clugston, S.L.; Barnard, J.F.; Kinach, R.; Miedema, D.; Ruman, R.; Daub, E.; Honek, J.F. Overproduction and characterization of a dimeric non-zinc glyoxalase I from Escherichia coli: Evidence for optimal activation by nickel ions. Biochemistry 1998, 37, 8754–8763. [Google Scholar] [CrossRef]
- Sukdeo, N.; Clugston, S.L.; Daub, E.; Honek, J.F. Distinct classes of glyoxalase I: Metal specificity of the Yersinia pestis, Pseudomonas aeruginosa and Neisseria meningitidis enzymes. Biochem. J. 2004, 384, 111–117. [Google Scholar] [CrossRef]
- Ozyamak, E.; Black, S.S.; Walker, C.A.; Maclean, M.J.; Bartlett, W.; Miller, S.; Booth, I.R. The critical role of S-lactoylglutathione formation during methylglyoxal detoxification in Escherichia coli. Mol. Microbiol. 2010, 78, 1577–1590. [Google Scholar] [CrossRef]
- Deponte, M. Glyoxalase diversity in parasitic protists. Biochem. Soc. Trans. 2014, 42, 473–478. [Google Scholar] [CrossRef]
- Youn, H.D.; Kim, E.J.; Roe, J.H.; Hah, Y.C.; Kang, S.O. A novel nickel-containing superoxide dismutase from Streptomyces spp. Biochem. J. 1996, 318 Pt 3, 889–896. [Google Scholar] [CrossRef]
- Dupont, C.; Neupane, K.; Shearer, J.; Palenik, B. Diversity, function and evolution of genes coding for putative Ni-containing superoxide dismutases. Environ. Microbiol. 2008, 10, 1831–1843. [Google Scholar] [CrossRef]
- Zambelli, B.; Ciurli, S. Nickel and human health. In Interrelations between Essential Metal Ions and Human Diseases; Springer: Dordrecht, The Netherlands, 2013; pp. 321–357. [Google Scholar]
- Lebrette, H.; Brochier-Armanet, C.; Zambelli, B.; de Reuse, H.; Borezee-Durant, E.; Ciurli, S.; Cavazza, C. Promiscuous nickel import in human pathogens: Structure, thermodynamics, and evolution of extracytoplasmic nickel-binding proteins. Structure 2014, 22, 1421–1432. [Google Scholar] [CrossRef]
- Chivers, P.T. Nickel recognition by bacterial importer proteins. Metallomics 2015, 7, 590–595. [Google Scholar] [CrossRef] [Green Version]
- Lebrette, H.; Iannello, M.; Fontecilla-Camps, J.C.; Cavazza, C. The binding mode of Ni-(L-His) 2 in NikA revealed by X-ray crystallography. J. Inorg. Biochem. 2013, 121, 16–18. [Google Scholar] [CrossRef]
- Ghssein, G.; Brutesco, C.; Ouerdane, L.; Fojcik, C.; Izaute, A.; Wang, S.; Hajjar, C.; Lobinski, R.; Lemaire, D.; Richaud, P. Biosynthesis of a broad-spectrum nicotianamine-like metallophore in Staphylococcus aureus. Science 2016, 352, 1105–1109. [Google Scholar] [CrossRef]
- Grim, K.P.; San Francisco, B.; Radin, J.N.; Brazel, E.B.; Kelliher, J.L.; Solórzano, P.K.P.; Kim, P.C.; McDevitt, C.A.; Kehl-Fie, T.E. The metallophore staphylopine enables Staphylococcus aureus to compete with the host for zinc and overcome nutritional immunity. MBio 2017, 8, e01281-17. [Google Scholar] [CrossRef]
- Song, L.; Zhang, Y.; Chen, W.; Gu, T.; Zhang, S.-Y.; Ji, Q. Mechanistic insights into staphylopine-mediated metal acquisition. Proc. Natl. Acad. Sci. USA 2018, 115, 3942–3947. [Google Scholar] [CrossRef] [Green Version]
- Lhospice, S.; Gomez, N.O.; Ouerdane, L.; Brutesco, C.; Ghssein, G.; Hajjar, C.; Liratni, A.; Wang, S.; Richaud, P.; Bleves, S. Pseudomonas aeruginosa zinc uptake in chelating environment is primarily mediated by the metallophore pseudopaline. Sci. Rep. 2017, 7, 17132. [Google Scholar] [CrossRef]
- Schauer, K.; Gouget, B.; Carrière, M.; Labigne, A.; De Reuse, H. Novel nickel transport mechanism across the bacterial outer membrane energized by the TonB/ExbB/ExbD machinery. Mol. Microbiol. 2007, 63, 1054–1068. [Google Scholar] [CrossRef]
- Zhang, P. Structure and mechanism of energy-coupling factor transporters. Trends Microbiol. 2013, 21, 652–659. [Google Scholar] [CrossRef]
- Bauerfeind, P.; Garner, R.M.; Mobley, L. Allelic exchange mutagenesis of nixA in Helicobacter pylori results in reduced nickel transport and urease activity. Infect. Immun. 1996, 64, 2877–2880. [Google Scholar]
- Fischer, F.; Robbe-Saule, M.; Turlin, E.; Mancuso, F.; Michel, V.; Richaud, P.; Veyrier, F.J.; De Reuse, H.; Vinella, D. Characterization in Helicobacter pylori of a nickel transporter essential for colonization that was acquired during evolution by gastric Helicobacter species. PLoS Pathog. 2016, 12, e1006018. [Google Scholar] [CrossRef]
- Nolan, K.J.; McGee, D.J.; Mitchell, H.M.; Kolesnikow, T.; Harro, J.M.; O’Rourke, J.; Wilson, J.E.; Danon, S.J.; Moss, N.D.; Mobley, H.L. In vivo behavior of a Helicobacter pylori SS1 nixA mutant with reduced urease activity. Infect. Immun. 2002, 70, 685–691. [Google Scholar] [CrossRef]
- Eitinger, T.; Suhr, J.; Moore, L.; Smith, J.A.C. Secondary transporters for nickel and cobalt ions: Theme and variations. Biometals 2005, 18, 399–405. [Google Scholar] [CrossRef]
- Hiron, A.; Posteraro, B.; Carriere, M.; Remy, L.; Delporte, C.; La Sorda, M.; Sanguinetti, M.; Juillard, V.; Borezee-Durant, E. A nickel ABC-transporter of Staphylococcus aureus is involved in urinary tract infection. Mol. Microbiol. 2010, 77, 1246–1260. [Google Scholar] [CrossRef]
- Remy, L.; Carrière, M.; Derré-Bobillot, A.; Martini, C.; Sanguinetti, M.; Borezée-Durant, E. The Staphylococcus aureus Opp1 ABC transporter imports nickel and cobalt in zinc-depleted conditions and contributes to virulence. Mol. Microbiol. 2013, 87, 730–743. [Google Scholar] [CrossRef]
- Lebrette, H.; Borezée-Durant, E.; Martin, L.; Richaud, P.; Erba, E.B.; Cavazza, C. Novel insights into nickel import in Staphylococcus aureus: The positive role of free histidine and structural characterization of a new thiazolidine-type nickel chelator. Metallomics 2015, 7, 613–621. [Google Scholar] [CrossRef]
- Chivers, P.T.; Benanti, E.L.; Heil-Chapdelaine, V.; Iwig, J.S.; Rowe, J.L. Identification of Ni-(L-His)2 as a substrate for NikABCDE-dependent nickel uptake in Escherichia coli. Metallomics 2012, 4, 1043–1050. [Google Scholar] [CrossRef]
- Subashchandrabose, S.; Hazen, T.H.; Brumbaugh, A.R.; Himpsl, S.D.; Smith, S.N.; Ernst, R.D.; Rasko, D.A.; Mobley, H.L. Host-specific induction of Escherichia coli fitness genes during human urinary tract infection. Proc. Natl. Acad. Sci. USA 2014, 111, 18327–18332. [Google Scholar] [CrossRef]
- Robinson, A.E.; Lowe, J.E.; Koh, E.I.; Henderson, J.P. Uropathogenic enterobacteria use the yersiniabactin metallophore system to acquire nickel. J. Biol. Chem. 2018, 293, 14953–14961. [Google Scholar] [CrossRef] [Green Version]
- Lamont, E.A.; Xu, W.W.; Sreevatsan, S. Host-Mycobacterium avium subsp. paratuberculosis interactome reveals a novel iron assimilation mechanism linked to nitric oxide stress during early infection. BMC Genom. 2013, 14, 694. [Google Scholar] [CrossRef]
- Stahler, F.N.; Odenbreit, S.; Haas, R.; Wilrich, J.; Van Vliet, A.H.; Kusters, J.G.; Kist, M.; Bereswill, S. The novel Helicobacter pylori CznABC metal efflux pump is required for cadmium, zinc, and nickel resistance, urease modulation, and gastric colonization. Infect. Immun. 2006, 74, 3845–3852. [Google Scholar] [CrossRef]
- Armbruster, C.E.; Forsyth-DeOrnellas, V.; Johnson, A.O.; Smith, S.N.; Zhao, L.; Wu, W.; Mobley, H.L.T. Genome-wide transposon mutagenesis of Proteus mirabilis: Essential genes, fitness factors for catheter-associated urinary tract infection, and the impact of polymicrobial infection on fitness requirements. PLoS Pathog. 2017, 13, e1006434. [Google Scholar] [CrossRef]
- De Reuse, H. Nickel and Virulence in Bacterial Pathogens. In The Biological Chemistry of Nickel; Zamble, D., Rowińska-Żyrek, M., Kozlowski, H., Eds.; The Royal Society of Chemistry: Cambridge, UK, 2017; pp. 339–356. [Google Scholar]
- Vinella, D.; Fischer, F.; Vorontsov, E.; Gallaud, J.; Malosse, C.; Michel, V.; Cavazza, C.; Robbe-Saule, M.; Richaud, P.; Chamot-Rooke, J.; et al. Evolution of Helicobacter: Acquisition by gastric species of two histidine-rich proteins essential for colonization. PLoS Pathog. 2015, 11, e1005312. [Google Scholar] [CrossRef]
- Ge, R.; Watt, R.M.; Sun, X.; Tanner, J.A.; He, Q.Y.; Huang, J.D.; Sun, H. Expression and characterization of a histidine-rich protein, Hpn: Potential for Ni2+ storage in Helicobacter pylori. Biochem. J. 2006, 393, 285–293. [Google Scholar] [CrossRef]
- Zeng, Y.-B.; Zhang, D.-M.; Li, H.; Sun, H. Binding of Ni2+ to a histidine-and glutamine-rich protein, Hpn-like. JBIC J. Biol. Inorg. Chem. 2008, 13, 1121. [Google Scholar] [CrossRef]
- Chiera, N.M.; Rowinska-Zyrek, M.; Wieczorek, R.; Guerrini, R.; Witkowska, D.; Remelli, M.; Kozlowski, H. Unexpected impact of the number of glutamine residues on metal complex stability. Metallomics 2013, 5, 214–221. [Google Scholar] [CrossRef] [Green Version]
- Seshadri, S.; Benoit, S.L.; Maier, R.J. Roles of His-rich hpn and hpn-like proteins in Helicobacter pylori nickel physiology. J. Bacteriol. 2007, 189, 4120–4126. [Google Scholar] [CrossRef]
- Saylor, Z.; Maier, R. Helicobacter pylori nickel storage proteins: Recognition and modulation of diverse metabolic targets. Microbiology 2018, 164, 1059–1068. [Google Scholar] [CrossRef]
- Benoit, S.L.; Miller, E.F.; Maier, R.J. Helicobacter pylori stores nickel to aid its host colonization. Infect. Immun. 2013, 81, 580–584. [Google Scholar] [CrossRef]
- Skouloubris, S.; Labigne, A.; De Reuse, H. The AmiE aliphatic amidase and AmiF formamidase of Helicobacter pylori: Natural evolution of two enzyme paralogues. Mol. Microbiol. 2001, 40, 596–609. [Google Scholar] [CrossRef]
- Saylor, Z.J. Helicobacter pylori nickel storage proteins: Recognition and modulation of diverse metabolic targets. Ph.D. Thesis, The University of Georgia, Athens, GA, USA, 2018. [Google Scholar]
- Lopez, A.J.; Martinez, L. Parametric models to compute tryptophan fluorescence wavelengths from classical protein simulations. J. Comput. Chem. 2018, 39, 1249–1258. [Google Scholar] [CrossRef]
- Deshayes, S.; Divita, G. Fluorescence technologies for monitoring interactions between biological molecules in vitro. Prog. Mol. Biol. Transl. Sci. 2013, 113, 109–143. [Google Scholar]
- Cvetkovic, A.; Menon, A.L.; Thorgersen, M.P.; Scott, J.W.; Poole, F.L., II; Jenney, F.E., Jr.; Lancaster, W.A.; Praissman, J.L.; Shanmukh, S.; Vaccaro, B.J. Microbial metalloproteomes are largely uncharacterized. Nature 2010, 466, 779. [Google Scholar] [CrossRef]
- Schauer, K.; Muller, C.; Carrière, M.; Labigne, A.; Cavazza, C.; De Reuse, H. The Helicobacter pylori GroES cochaperonin HspA functions as a specialized nickel chaperone and sequestration protein through its unique C-terminal extension. J. Bacteriol. 2010, 192, 1231–1237. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, J.; Yang, F.; Wu, W.; Sun, H.; Xie, Q.; Si, W.; Zou, Q.; Yang, Z. Immunization with heat shock protein A and gamma-glutamyl transpeptidase induces reduction on the Helicobacter pylori colonization in mice. PLoS ONE 2015, 10, e0130391. [Google Scholar]
- Zhang, X.J.; Feng, S.Y.; Li, Z.T.; Feng, Y.M. Expression of Helicobacter pylori hspA Gene in Lactococcus lactis NICE System and Experimental Study on Its Immunoreactivity. Gastroenterol. Res. Pract. 2015, 2015, 750932. [Google Scholar] [CrossRef]


| Pathogen | Ni-enzyme * | Role in Pathogenesis (Reference) |
|---|---|---|
| EUKARYOTES | ||
| Human fungi | ||
| Cryptococcus neoformans | Ure | Virulence factor in experimental cryptococcosis [17] |
| Required for microvascular sequestration and mouse brain invasion [18] | ||
| Modulates phagolysosomal pH; important for mouse brain infection [19] | ||
| Released via extracellular vesicles [20] | ||
| Cryptococcus gattii | Ure | Virulence factor in mice [21] |
| Coccidioides posadasii | Ure | Coccidioidomycosis in mice [22,23] |
| Histoplasma capsulatum | Ure | Released via extracellular vesicles [24] |
| Paracoccidioides brasiliensis | Ure | Up-expressed in mouse infection model [25] |
| Oomycetes | ||
| Pythium insidiosum | Ure | Putative virulence factor for pythiosis [26] |
| Protists | ||
| Leishmania major | Glo-I | Important for parasite metabolism: methylglyoxal detoxification [27] |
| Leishmania donovani | Glo-I | Essential for growth; suggested as drug target [28] |
| Trypanosoma cruzi | Ard | Important for parasite metabolism: methionine salvage pathway |
| Glo-I | Important for parasite metabolism: methylglyoxal detoxification [29] | |
| PROKARYOTES | ||
| Actinobacteria | ||
| Actinomyces naeslundii | Ure | Needed in acidic environment; promotes plaque formation [30] |
| Corynebacterium urealyticum | Ure | Plays an important role in urinary tract infection [31] |
| Mycobacterium tuberculosis | Hyc | Essential for optimal growth [32] |
| Up-regulated during infection of human macrophage-like cells [33] | ||
| Up-expressed in resting and active murine bone marrow macrophages [34] | ||
| Ure | Important for survival under nitrogen-limited environment [35] | |
| Streptomyces scabies | Sod | Important against oxidative stress encountered in host (plant) |
| Firmicutes | ||
| Clostridia | Glo-I | Important for metabolism: methylglyoxal detoxification [36] |
| Staphylococcus aureus | Ure | Increased expression of structural and accessory genes in biofilms [37] |
| Required for acid response and persistent murine kidney infection [38] | ||
| Decreased activity in mixed source (S. epidermidis) biofilms [39] | ||
| Staphylococcus epidermidis | Ure | Decreased activity in mixed source (S. aureus) biofilms [39] |
| Staphylococcus saprophyticus | Ure | Important for bladder infection and bladder stones in rats [40] |
| Streptococcus salivarius | Ure | Important as source of nitrogen and to combat acid stress [41] |
| Mollicutes | ||
| Ureaplasma urealyticum | Ure | Role in human vaginal infection; used for diagnostic [42] |
| Ammonia contributes to PMF-driven ATP synthesis [43] | ||
| Ammonia generates struvite stone formation in rat bladders [44] | ||
| Ureaplasma parvum | Ure | Role in human vaginal infection; used for diagnostic [42] |
| Ureaplasma diversum | Ure | Role in vaginal infection of cattle and small ruminants [45] |
| Proteobacteria | ||
| Alphaproteobacteria | ||
| Brucella abortus | Ure | Needed for intestinal colonization in a murine model [46] |
| Immunization with B. a. urease protects against B. abortus infection in mice [47] | ||
| Brucella melitensis | Ure | Immunization with B. a. urease protects against B. melitensis in mice [47] |
| Brucella suis | Ure | Required for intestinal colonization in a murine model [48] |
| Immunization with B. a. urease protects against B. suis infection in mice [47] | ||
| Betaproteobacteria | ||
| Neisseria meningitides | Glo-I | Important for methylglyoxal detoxification and potassium efflux (hypothesized) |
| Neisseria gonorrhoeae | Glo-I | Important for methylglyoxal detoxification and potassium efflux (hypothesized) |
| Gammaproteobacteria | ||
| All γ-proteobacteria | Ard | Important for metabolism: methionine salvage pathway |
| All γ-proteobacteria | Glo-I | Important for methylglyoxal detoxification, potassium efflux |
| Acinetobacter baumannii | Ure | Virulence factor in worm and amoeba hosts [49] |
| Acinetobacter lwoffii | Ure | Needed to survive in the stomach [50] |
| Actinobacillus | Ure | Important for swine respiratory tract infection [51,52] |
| pleuropneumoniae | Hyd-1 | Important for (PMF-driven) metabolism and motility |
| Escherichia coli | Hyd-2 | Important for (PMF-driven) metabolism and motility |
| Hyc | Needed (as part of FHL) to dissipate formic acid-induced acidity [53] | |
| E. coli (Shiga-toxin producing) | Ure | Needed for colonization in the murine model [54] |
| Edwardsiella tarda | Hyd | Hyd. accessory protein Sip2 essential for acid resistance and host infection [55] |
| Haemophilus influenzae | Ure | Important for acid resistance, expressed during human pulmonary infection [56] |
| Klebsiella pneumoniae | Ure | Required for colonization in murine intestinal model [57] |
| Morganella morganii | Ure | Needed for survival at low pH [58,59] |
| Proteus mirabilis | Hyd | Important for swarming motility [60] |
| Ure | Role in persistence, urolithiasis, and acute pyelonephritis in a mouse model [61] | |
| Role in extracellular crystal stone cluster formation in the bladder [62] | ||
| Induced in polymicrobial biofilms [63] | ||
| Providencia stuartii | Ure | Involved in crystal stones formation; induced in polymicrobial biofilms [59] |
| Pseudomonas aeruginosa | Glo-I | Important for methylglyoxal detoxification and potassium efflux (hypothesized) |
| Salmonella Typhimurium | Hyd-1 | Important for acid resistance and macrophage colonization [64] |
| Hyd-2 | Most important hydrogenase for gut invasion [65,66] | |
| Hyd-5 | Expressed under aerobic conditions and in macrophages [64,67,68] | |
| Hyc | Important for anaerobic acid resistance [69] | |
| Shigella flexneri | Hyd | Important for acid resistance [70] |
| Vibrio parahaemolyticus | Ure | Important for pathogenicity [71] |
| Yersinia enterocolitica | Ure | Important for survival at low pH [58,72] |
| Yersinia pestis | Glo-I | Important for methylglyoxal detoxification and potassium efflux (hypothesized) |
| Deltaproteobacteria | ||
| Bilophila wadsworthia | Hyd | H2 used as energy source, optimal growth in presence of H2 and taurine [73] |
| Epsilonproteobacteria | ||
| Campylobacter jejuni | Hyd | Important for chicken cecum colonization [74] |
| Essential for chicken colonization in absence of formate dehydrogenase [74] | ||
| Required for in vitro interaction with human intestinal cells [75] | ||
| Campylobacter concisus | Hyd | Essential for growth under microaerobic conditions [76] |
| Helicobacter hepaticus | Hyd | Role in amino-acid transport and causing liver lesions in mice [77] |
| Ure | Promotes hepatic inflammation in mice [78] | |
| Helicobacter mustelae | Ure | Essential for ferret stomach colonization [79] |
| Helicobacter pylori | Hyd | Needed for mouse stomach colonization [80] |
| Role in CO2 fixation [81] | ||
| Role in CagA translocation [82] | ||
| Ure | Cytotoxic effect on Caco-2 cells [83] | |
| Needed for nude mouse stomach colonization [84] | ||
| Essential for gnotobiotic piglet stomach colonization [85] | ||
| Activates human phagocytes and macrophages [86,87] | ||
| Binds to class II MHC on gastric epithelial cells and induces their apoptosis [88] | ||
| Essential for Mongolian gerbil colonization [89,90] | ||
| Urease-produced CO2 protects against host peroxynitrite [91] | ||
| Urease-produced ammonia disrupts tight cell junction integrity [92] | ||
| Dysregulates epithelial tight junctions through myosin activation [93] | ||
| Activates blood platelets through a lipoxygenase-mediated pathway [94] | ||
| Alters mucin gene expression in human gastric cells [95] | ||
| Essential for chronic mouse infection [96] | ||
| Role in angiogenesis, endothelial cells and chicken embryo models [97] | ||
| Induces blood platelets inflammatory pathways [98] | ||
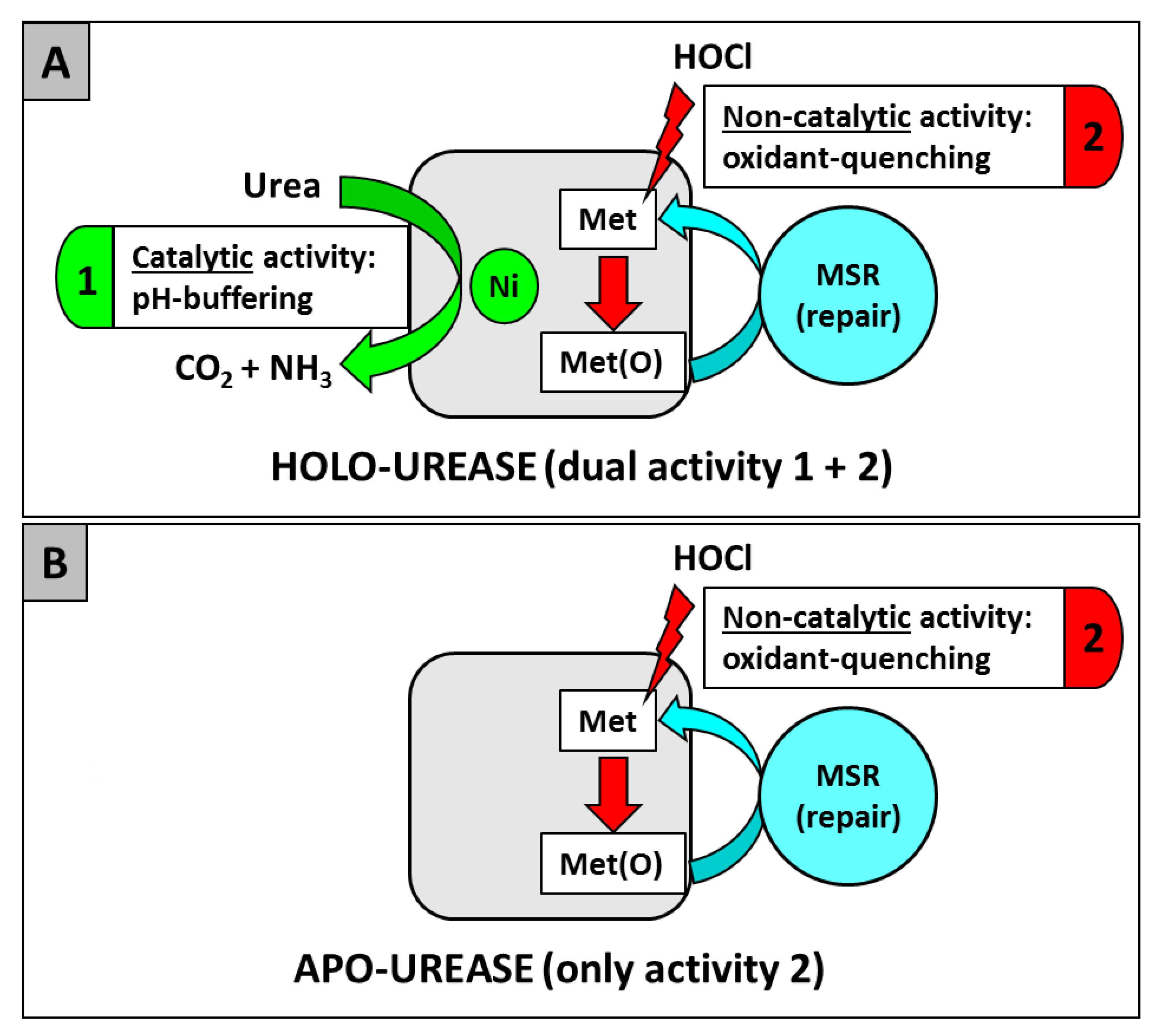
| Non catalytic, oxidative stress-combatting role [99] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maier, R.J.; Benoit, S.L. Role of Nickel in Microbial Pathogenesis. Inorganics 2019, 7, 80. https://0-doi-org.brum.beds.ac.uk/10.3390/inorganics7070080
Maier RJ, Benoit SL. Role of Nickel in Microbial Pathogenesis. Inorganics. 2019; 7(7):80. https://0-doi-org.brum.beds.ac.uk/10.3390/inorganics7070080
Chicago/Turabian StyleMaier, Robert J., and Stéphane L. Benoit. 2019. "Role of Nickel in Microbial Pathogenesis" Inorganics 7, no. 7: 80. https://0-doi-org.brum.beds.ac.uk/10.3390/inorganics7070080




