Recent Strategies for the Remediation of Textile Dyes from Wastewater: A Systematic Review
Abstract
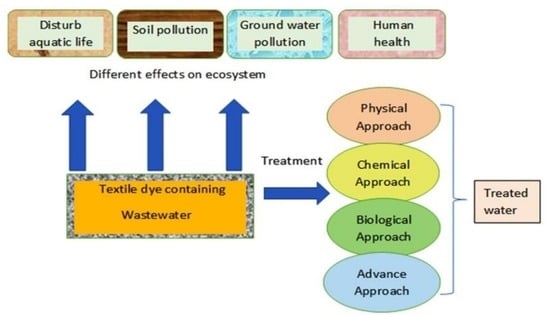
:1. Introduction
2. Data Collection and Bibliometric Analyses
3. Dyes Used in Textile Industry
4. Approaches for Dyes Remediation
4.1. Physical and Chemical Approaches
4.1.1. Adsorption
4.1.2. Ion-Exchange Method
4.1.3. Membrane Filtration
4.1.4. Fenton Process
4.1.5. Ozonation
4.2. Biological Approaches
4.2.1. Enzymatic Method
4.2.2. Microbial Remediation
Bacterial Remediation
Pure Bacterial Cultures
Mixed Bacterial Cultures
Actinomycetes
Phycoremediation
Yeast-Mediated Dye Decolorization
Phytoremediation
5. Recent Strategies for Remediation of Dyes
5.1. Genetically Engineered Microorganisms (GEMs)
5.2. Microbial Biosorbents
5.3. Bioreactors
5.4. Nanoparticles Based Bioremediation
5.5. Microbial Fuel Cells (MFCs)
6. Challenges and Future Perspectives
7. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Petersen, L.; Heynen, M.; Pellicciotti, F. Freshwater resources; past, present, future. Int. Encycl. Geogr. People Earth Environ. Technol. 2016, 1–11. [Google Scholar] [CrossRef]
- Younis, S.A.; Serp, P.S.; Nassar, H.N. Photocatalytic and biocidal activities of ZnTiO2 oxynitrite heterojunction with MOF-5 and G-C3N4: A case study for textile wastewater treatment under direct sunlight. J. Hazard. Mater. 2021, 410, 124562. [Google Scholar] [CrossRef]
- Akpomie, K.G.; Conradie, J. Advances in application of cotton-based adsorbents for heavy metalstrapping, surface modifications and future perspectives. Ecotoxicol. Environ. Saf. 2020, 201, 110825. [Google Scholar] [PubMed]
- Al-Tohamy, R.; Ali, S.S.; Li, F.; Okasha, K.M.; Mahmoud, Y.A.G.; Elsamahy, T.; Jiao, H.; Fu, Y.; Sun, J. A critical review on the treatment of dye-containing wastewater: Ecotoxicological and health concerns of textile dyes and possible remediation approaches for environmental safety. Ecotoxicol. Environ. Saf. 2022, 231, 113160. [Google Scholar] [CrossRef] [PubMed]
- Kishor, R.; Purchase, D.; Saratale, G.D.; Saratale, R.G.; Ferreira, L.F.R.; Bilal, M.; Chandra, R.; Bharagava, R.N. Ecotoxicological and health concerns of persistent coloring pollutants of textile industry wastewater and treatment approaches for environmental safety. Environ. Chem. Eng. 2021, 9, 105012. [Google Scholar]
- Garg, S.K.; Tripathi, M. Microbial strategies for discoloration and detoxification of azo dyes from textile effluents. Res. J. Microb. 2017, 12, 1–19. [Google Scholar]
- Parmar, S.; Daki, S.; Bhattacharya, S.; Shrivastav, A. Microorganism: An ecofriendly tool for waste management and environmental safety. In Development in Wastewater Treatment Research and Processes; Elsevier: Amsterdam, The Netherlands, 2022; pp. 175–193. [Google Scholar]
- Sharma, K.; Sharma, P.; Dhiman, S.K.; Chadha, P.; Saini, H.S. Biochemical, genotoxic, histological and ultrastructural effects on liver and gills of fresh water fish Channa punctatus exposed to textile industry intermediate 2 ABS. Chemosphere 2022, 287, 132103. [Google Scholar]
- Xu, M.; Guo, J.; Suh, G. Biodegradation of textile azo dye by Shewanella decolorationis S12 under micro aerophillic conditions. Appl. Microbiol. Biotechnol. 2007, 76, 719–726. [Google Scholar] [CrossRef]
- Tripathi, M.; Pathak, N.; Chaudhary, V.K.; Singh, P.; Singh, P.K.; Thirumalesh, B.V.; Bala, S.; Maurya, A.K.; Patel, N.; Yadav, B.K. Microbial decolorization of crystal violet dye by a native multimetal tolerant Aeromonas caviae MT-1 isolate from dye contaminated soil: Optimization and phototoxicity study. Toxicol. Int. 2023, 30, 83–93. [Google Scholar] [CrossRef]
- Vankar, P.S. Handbook on Natural Dyes for Industrial Applications: Extraction of Dyestuff from Flowers, Leaves, Vegetables; Niir Project Consultancy Services: Delhi, India, 2016. [Google Scholar]
- Elgarahy, A.M.; Elwakeel, K.Z.; Mohammad, S.H.; Elshoubaky, G.A. A critical review of biosorption of dyes, heavy metals and metalloids from wastewater as an efficient and green process. Clean. Eng. Technol. 2021, 4, 100209. [Google Scholar]
- Tripathi, M.; Kumar, S.; Singh, D.N.; Pandey, R.; Pathak, N.; Fatima, H. Bioremediation of dye contaminated soil. In Soil Bioremediation: An Approach towards Sustainable Technology; Parray, J.A., Hashem, A., Mahmoud, A.E., Eds.; John Wiley and Sons: Hoboken, NJ, USA, 2021; pp. 115–142. [Google Scholar]
- Tripathi, M.; Singh, P.; Singh, R.; Bala, S.; Pathak, N.; Singh, S.; Chauhan, R.S.; Singh, P.K. Microbial biosorbent for remediation of dyes and heavy metal pollution: A green strategy for sustainable environment. Front. Microbiol. 2023, 14, 1168954. [Google Scholar] [CrossRef] [PubMed]
- Sharma, B.; Dangi, A.K.; Shukla, P. Contemporary enzyme based technologies for bioremediation: A review. J. Environ. Manag. 2018, 210, 10–22. [Google Scholar] [CrossRef] [PubMed]
- Muthu, S.S. Sustainability in the Textile Industry; Springer: Berlin/Heidelberg, Germany, 2017; pp. 1–8. [Google Scholar]
- Khan, S.; Malik, A. Toxicity evaluation of textile effluents and role of native soil bacterium in biodegradation of a textile dye. Environ. Sci. Pollut. Res.-Int. 2018, 25, 4446–4458. [Google Scholar] [CrossRef] [PubMed]
- Seo, Y.H.; Park, D.; Oh, Y.K.; Yoon, S.; Han, J.I. Harvesting of microalgae cell using oxidised dye wastewater. Bioresour. Technol. 2015, 192, 802–806. [Google Scholar] [CrossRef]
- Goswami, M.; Chaturvedi, P.; Sonwani, R.K. Application of arjuna (Terminalia arjuna) seed biochor in hybrid treatment system for the bioremediation of congo red dye. Bioresour. Technol. 2020, 307, 123203. [Google Scholar] [CrossRef]
- Kiran, S.; Ashraf, A.; Rahmat, M. Green synthesis of magnesium oxide nanoparticles using leaves of Iresine herbstii for remediation of reactive brown 9 dye. Glob. NEST J. 2022, 24, 291–296. [Google Scholar]
- Sathishkumar, K.; Al Salhi, M.S.; Sanganyado, E.; Devanesan, S.; Arulprakash, A.; Rajasekar, A. Sequential electrochemical oxidation and bio treatment of the azo dye congo red and textile effluent. J. Photochem. Photobiol. B 2019, 200, 111655. [Google Scholar] [CrossRef]
- Shetty, K.; Krishnakumar, G. Algal and cyanobacterial biomass as potential dye biodecolorizing material: A review. Biotechnol. Lett. 2020, 42, 2467–2488. [Google Scholar] [CrossRef]
- Prabhakar, Y.; Gupta, A.; Kaushik, A. Using indigenous bacteria isolate Nesterenkonia lacusekhoensis for removal of azo dyes: A low cost friendly approach for bioremediation of textile wastewaters. Environ. Dev. Sustain. 2022, 24, 5344–5367. [Google Scholar] [CrossRef]
- Zhao, J.; Dang, Z.; Muddassir, M.; Raza, S.; Zhong, A.; Wang, X.; Jin, J. A new Cd(II)-based coordination polymer for efficient photocatalytic removal of organic dyes. Molecules 2023, 28, 6848. [Google Scholar] [CrossRef]
- Zheng, M.; Chen, J.; Zhang, L.; Cheng, Y.; Lu, C.; Liu, Y.; Singh, A.; Trivedi, M.; Kumar, A.; Liu, J. Metal organic frameworks as efficient adsorbents for drugs from wastewater. Mater. Today Commun. 2022, 31, 103514. [Google Scholar] [CrossRef]
- Zhong, Y.; Chen, C.; Liu, S.; Lu, C.; Liu, D.; Pan, Y.; Sakiyama, H.; Muddassir, M.; Liu, J. A new magnetic adsorbent of eggshell-zeolitic imidazolate framework for highly efficient removal of norfloxacin. Dalton Trans. 2021, 50, 18016–18026. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Lu, L.; Liu, D.; Wang, J.; Sakiyama, H.; Muddassir, M.; Nezamzadeh-Ejhieh, A.; Liu, J. Series of highly stable Cd(ii)-based MOFs as sensitive and selective sensors for detection of nitrofuran antibiotic. CrystEngComm 2021, 23, 8043–8052. [Google Scholar] [CrossRef]
- Bala, S.; Garg, D.; Thirumalesh, B.V.; Sharma, M.; Sridhar, K.; Inbaraj, B.S.; Tripathi, M. Recent strategies for bioremediation of emerging pollutants; a review for a green and sustainable environment. Toxics 2022, 10, 484. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Quanungo, K. Challenges in effluents treatment containing dye. Adv. Res. Text. Eng. 2022, 7, 1075. [Google Scholar]
- Tripathi, M.; Singh, D.N. Bioremediation: Challenges and Advancements; Bentham Science Publishers: Singapore, 2022. [Google Scholar]
- Benkhaya, S.; M’rabet, S.; El Harfi, A. Classifications, properties, recent synthesis and applications of azo dyes. Heliyon 2020, 6, e03271. [Google Scholar] [CrossRef]
- Benkhaya, S.; El Harfi, S.; El Harfi, A. Classifications, properties and applications of textile dyes: A review. Appl. J. Environ. Eng. Sci. 2017, 3, 311–320. [Google Scholar]
- Dhodapkar, R.; Rao, N.N.; Pande, S.P.; Kaul, S.N. Removal of basic dyes from aqueous medium using a novel polymer. Jalshakti Bioresour. Technol. 2006, 97, 877–885. [Google Scholar] [CrossRef]
- Yavuz, O.; Aydin, A.H. Removal of direct dyes from aqueous solution using various adsorbent. Pol. J. Environ. Stud. 2006, 15, 155–161. [Google Scholar]
- Bozic, M.; Kokol, V. Ecological alternatives to the reduction and oxidation processes in dyeing with vat and sulfur dyes. Dyes Pigment. 2008, 76, 299–309. [Google Scholar] [CrossRef]
- Holkar, C.R.; Jadhav, A.J.; Pinjari, D.V.; Mahamuni, N.M.; Pandit, A.B. A critical review on textile wastewater treatment: Possible approaches. J. Environ. Manag. 2016, 182, 351–366. [Google Scholar] [CrossRef] [PubMed]
- Mudhoo, A.; Ramasamy, D.L.; Bhatnagar, A.A.; Usman, M.; Sillanpaa, M. An analysis of the versatility and effectiveness of composts for sequestering heavy metal ions, dyes and xenobiotics from soils and aqueous milieus. Ecotoxicol. Environ. Saf. 2020, 197, 110587. [Google Scholar] [CrossRef] [PubMed]
- Samsami, S.; Mohamadizaniani, M.; Sarrafzadeh, M.H.; Rene, E.R.; Firoozbahr, M. Recent advances in the treatment of dye containing wastewater from textile industries: Overview and perspectives. Process Saf. Environ. Prot. 2020, 143, 138–163. [Google Scholar] [CrossRef]
- Abu-Nada, A.; Abdala, A.; McKay, G. Removal of phenols and dyes from aqueous solutions using graphene and graphene composite adsorption; a review. J. Environ. Chem. Eng. 2021, 9, 105858. [Google Scholar] [CrossRef]
- Jadhav, A.C.; Jadhav, N.C. Treatment of textile wastewater using adsorption and adsorbents. In Suitable Technologies for Textile Wastewater Treatments; Woodhead Publishing: Sawston, UK, 2021; pp. 235–373. [Google Scholar]
- Hisada, M.; Tomizawa, Y.; Kaware, Y. Removal kinetics of cationic azo-dye from aqueous solution by gamma- glutamic acid biosorbent contribution of adsorption and complexation/precipitation to basic orange 2 removal. J. Environ. Chem. Eng. 2019, 7, 103157. [Google Scholar] [CrossRef]
- Kubra, K.T.; Sulman, M.S.; Haisan, M.N. Enhanced toxic dye removal from wastewater using biodegradable polymeric natural adsorbent. J. Mol. Liquids 2021, 328, 115468. [Google Scholar] [CrossRef]
- Saifi, A.; Joseph, J.P.; Singh, A.P.; Pal, A.; Kumar, K. Complexation of an azo dye by cyclodextrins: A potential strategy for water purification. ACS Omega 2021, 6, 4776–4782. [Google Scholar] [CrossRef]
- Briao, G.V.; Jahn, S.L.; Foletto, E.L.; Dotto, G.L. Highly efficient and reusable mesoporous zeolite synthesized from a biopolymer for cationic dyes adsorption. Colloids Surf. A Physicochem. Eng. Asp. 2018, 556, 43–50. [Google Scholar] [CrossRef]
- Madan, S.; Shaw, R.; Tiwari, S.; Tiwari, S.K. Adsorption dynamics of congo red dye removal using ZnO2 functionalized high silica zeolite particles. J. Appl. Surf. Sci. 2019, 187, 907–917. [Google Scholar] [CrossRef]
- Harja, M.; Buema, G.; Bucur, D. Recent advances in removal of Congo Red dye by adsorption using an industrial waste. Sci. Rep. 2022, 12, 6087. [Google Scholar] [CrossRef]
- Minisy, I.M.; Salahuddin, N.A.; Ayad, M.M. Chitosan/polyaniline hybrid for the removal of cationic and anionic dyes from aqueous solutions. J. Appl. Polym. Sci. 2019, 136, 47056. [Google Scholar] [CrossRef]
- Bober, P.; Minisy, I.M.; Acharya, U.; Pfleger, J.; Babayan, V.; Kazantseva, N.; Hodan, J.; Stejskal, J. Conducting polymer composite aerogel with magnetic properties for organic dye removal. Synth. Met. 2020, 260, 116266. [Google Scholar] [CrossRef]
- Ayad, M.M.; Amer, W.A.; Zaghlol, S.; Minisy, I.M.; Bober, P.; Stejskal, J. Polypyrrole-coated cotton textile as adsorbent of methylene blue dye. Chem. Pap. 2018, 72, 1605–1618. [Google Scholar] [CrossRef]
- Ahmad, A.; Setapar, M.S.H.; Chuong, C.S.; Khatoon, A.; Wani, W.A.; Kumar, R.; Rafatullah, M. Recent advances in new generation dye removal technologies; novel search for approaches to reprocess wastewater. RSC Adv. 2015, 5, 30801–30818. [Google Scholar] [CrossRef]
- Xu, T. Ion exchange membranes: State of this development and perspective. Memb. Sci. 2005, 263, 1–29. [Google Scholar] [CrossRef]
- Marin, N.M.; Pascu, L.F.; Demha, A.; Nita-Lazar, M.; Badea, L.; Aboul-Enein, H.Y. Removal of the acid orange 10 by an ion-exchange and microbiological methods. Int. J. Environ. Sci. Technol. 2019, 16, 6357–6366. [Google Scholar] [CrossRef]
- Waly, A.I.; Khedr, M.A.; Ali, H.M.; Riad, B.Y.; Ahmed, I.M. Synthesis and characterization of ion- exchange based on waste cotton for dye removal from wastewater. Egypt. J. Chem. 2019, 62, 451–468. [Google Scholar]
- Bayramoglu, G.; Kunduzcu, G.; Arica, M.Y. Prepartion and characterization of strong cation exchange ter polymer resin as effective adsorbent for removal of dispersed dyes. Polym. Eng. Sci. 2020, 60, 192–201. [Google Scholar] [CrossRef]
- Zheng, Y.; Yao, G.; Cheng, Q.; Yu, S.; Liu, M.; Gao, C. Positively charged thin–film composite hollow fibre nanofiltration membrane for the removal of cationic dyes through submerged filtration. Desalination 2013, 328, 42–50. [Google Scholar] [CrossRef]
- Rambabu, K.; Bharath, G.; Monash, P.; Velu, S.; Banat, F.; Naushad, M.; Arthanareeswaran, G.; Show, P.L. Effective treatment of dye polluted wastewater using nanoporous CaCl2 modified polyethersulfone membrane. Process Saf. Environ. Prot. 2019, 124, 266–278. [Google Scholar] [CrossRef]
- Cherayan, M. Ultrafiltration and Microfiltration Handbook; CRC Press: Boca Raton, FL, USA, 1998. [Google Scholar]
- Collivignarelli, M.C.; Abba, A.; Carnovale, M.; Damiani, S. Treatments for color removal from wastewater: State of the art. J. Environ. Manag. 2019, 236, 727–745. [Google Scholar] [CrossRef] [PubMed]
- Alventosa-deLara, E.; Barredo-Damas, S.; Alcaina-Miranda, M.I.; Iborasa-Ciar, M.I. Ultrafiltration technology with a ceramic membrane for reactive dye removal; optimization of membrane performance. J. Hazard. Mater. 2012, 209–210, 492–500. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.J.; Wang, H.C.; Cheng, H.Y.; Liang, B.; Liu, W.Z.; Han, J.L.; Zhang, B.; Wang, S.S. Electrochemistry stimulated environmental bioremediation development of applicable modular electrode and system scale up. Environ. Sci. Ecotechnol. 2020, 3, 100050. [Google Scholar]
- Wang, X.; Xia, J.; Ding, S.; Zhang, S.; Li, M.; Shang, Z.; Lu, J.; Ding, J. Removing organic matters from reverse osmosis concentrate using advanced oxidation biological activated carbon process combined with Fe3+/humus–reducing bacteria. Ecotoxicol. Environ. Saf. 2020, 203, 110945. [Google Scholar] [CrossRef]
- Lu, Z.; Hu, F.; Li, H.; Zhang, X.; Yu, S.; Liu, M.; Gao, C. Composite nano-filtration membrane with asymmetric selective separation layer for enhanced separation efficiency to anionic dye aqueous solution. J. Hazard. Mater. 2019, 368, 436–443. [Google Scholar] [PubMed]
- Shao, H.; Qi, Y.; Liang, S.; Qin, S.; Yu, J. Polypropylene composite hollow fibre ultrafiltration membranes with an acrylic hydrogel surface by in-situ ultrasonic wave assisted polymerization for dye removal. J. Appl. Polym. Sci. 2019, 136, 47099. [Google Scholar] [CrossRef]
- Fradj, A.B.; Boubakri, A.; Hafiane, A.; Hamouda, S.B. Removal of azoic dyes from aqueous solutions by chitosan enhanced ultrafilteration. J. Result Chem. 2020, 2, 100017. [Google Scholar] [CrossRef]
- Xiang, H.; Ren, G.; Yang, X.; Xu, D.; Zhang, Z.; Wang, X. A low cost solvent free method to synthesize alpha Fe2O3 nanoparticles with applications to degrade methyl orange in photofenton system. Ecotoxicol. Environ. Saf. 2020, 200, 110744. [Google Scholar] [CrossRef]
- Zhong, J.; Yang, B.; Feng, Y.; Chen, Y.; Wang, L.-G.; You, W.-D.; Ying, G.-G. Enhanced Photo–Fenton Removal Efficiency with Core-Shell Magnetic Resin Catalyst for Textile Dyeing Wastewater Treatment. Water 2021, 13, 968. [Google Scholar] [CrossRef]
- Saratale, R.G.; Sivapathan, S.; Saratale, G.D.; Banu, J.R.; Kim, D.S. Hydroxamic acid mediated heterogenous fenton like catalyst for efficient removal of acid red 88, textile wastewater and their phytotoxicity studies. Ecotoxicol. Environ. Saf. 2019, 167, 385–395. [Google Scholar] [CrossRef]
- Parsons, S. Advanced Oxidation Processes for Water Wastewater Treatment; IWA Publishing: London, UK, 2004. [Google Scholar]
- Gogate, P.R.; Pandit, A.B. A review of imperative technologies for wastewater treatment 1: Oxidation technologies at ambient conditions. J. Adv. Environ. Resour. 2004, 8, 501–551. [Google Scholar] [CrossRef]
- Ledakowicz, S.; Maciejewska, R.; Gebicka, L.; Perkowski, J. Kinetics of decolourization by Fenton’s reagent. J. Ozone Sci. Eng. 2000, 22, 195–205. [Google Scholar] [CrossRef]
- Chen, Y.; Cheng, Y.; Guan, X.; Liu, Y.; Nie, J.; Li, C. A rapid fenton treatment of bio-treated dyeing and finishing wastewater at second scale intervals; kinetics by stopped-flow technique and application in a full scale plan. Sci. Rep. 2019, 9, 9689. [Google Scholar] [CrossRef] [PubMed]
- Zawadzki, P.; Deska, M. Degradation efficiency and kinetics analysis of an advance oxidation process, utilizing ozone, hydrogen peroxide and persulfate to degrade the dye rhodamine. Catalysts 2021, 11, 974. [Google Scholar] [CrossRef]
- Shriram, B.; Kanmani, S. Ozonation of Textile Dyeing Wastewater: A Review. 2014, 2014-15, p. 3. Available online: https://www.researchgate.net/profile/ShriramBalasubramanyan/publication/281776011_Ozonation_of_Textile_Dyeing_Wastewater_-_A_Review/links/55f7f43e08ae07629dcded7a/Ozonation-of-Textile-Dyeing-Wastewater-A-Review.pdf (accessed on 16 November 2023).
- Venkatesh, S.; Venkatesh, K.; Quaff, A.R. Dye decomposition by combined ozonation and anaerobic treatment: Cost effective technology. J. Appl. Res. Technol. 2017, 15, 340–345. [Google Scholar] [CrossRef]
- Cardoso, R.M.F.; Cardoso, I.M.F.; da Silva, P.L.; Joaguim, C.G.; da Silva, E. Copper (2) doped carbon dots as catalyst for ozone degradation of textile dyes. Nanomaterials 2022, 12, 1211. [Google Scholar] [CrossRef]
- Lanzetta, A.; Papirio, S.; Oliva, A.; Cesaro, A.; Pucci, L.; Capasso, E.M.; Esposito, G.; Pirozzi, F. Ozonation processes for color Removal from Urban and Leather Tanning Wastewater. Water 2023, 15, 2362. [Google Scholar] [CrossRef]
- Chen, K.C.; Wu, J.Y.; Liou, D.J.; Hwang, S.C.J. Decolorization of the textile dye by newly isolated bacterial strains. J. Biotechnol. 2003, 101, 57–68. [Google Scholar] [CrossRef]
- Patel, H.; Yadav, V.K.; Yadav, K.K.; Choudhary, N.; Kalasariya, H.; Alam, M.M.; Gacem, A.; Amanullah, M.; Ibrahium, H.A.; Park, J.W.; et al. A review: A recent and systematic approach towards microbial biodegradation of dyes from textile industries. Water 2022, 14, 3163. [Google Scholar] [CrossRef]
- Das, S.; Cherwoo, L.; Singh, R. Decoding dye degradation: Microbial remediation of textile industry effluents. Biotechnol. Notes 2023, 4, 64–76. [Google Scholar] [CrossRef]
- Pourbabaee, A.A.; Malakzadeh, F.; Sarbolouki, M.N.; Najafi, F. Aerobic decolorization and detoxification of disperse dye in textile effluent by a new isolate of Bacillus spp. Biotechnol. Bioeng. 2006, 93, 631–635. [Google Scholar] [CrossRef] [PubMed]
- Bhandari, S.; Poudel, D.K.; Marahatha, R.; Dawadi, S.; Khadaya, K.; Phuyal, S. Microbial enzymes used in bioremediation. J. Chem. 2021, 2021, 8849512. [Google Scholar] [CrossRef]
- Pham, V.H.T.; Kim, J.; Chang, S.; Bang, D. Investigating bio-inspired degradation of toxic dyes using potential multi-enzyme producing extremophiles. Microorganisms 2023, 11, 1273. [Google Scholar] [CrossRef] [PubMed]
- Mendes, S.; Robalo, M.P.; Martins, L.O. Bacterial enzymes and multi-enzymatic systems for cleaning-up dyes from the environment. In Microbial Degradation of Synthetic Dyes in Wastewater; Springer International Publishing: Berlin/Heidelberg, Germany, 2015; pp. 27–55. [Google Scholar]
- Chandra, R. Environmental Waste Management; CRC Press: Boca Raton, FL, USA, 2016. [Google Scholar]
- Hussain, Q. Peroxidase mediated decolourization and remediation of wastewater containing industrial dyes: A review. Rev. Environ. Sci. Biotechnol. 2010, 9, 117–140. [Google Scholar] [CrossRef]
- Lellis, B.; Polonio, C.Z.F.; Pamphile, J.A.; Polonio, J.C. Effects of textile dyes on health and the environment and bioremediation potential of living organisms. Biotechnol. Res. Innov. 2019, 3, 275–290. [Google Scholar] [CrossRef]
- Pinheiro, L.R.S.; Gradíssimo, D.G.; Xavier, L.P.; Santos, A.V. Degradation of azo dyes: Bacterial potential for bioremediation. Sustainability 2022, 14, 1510. [Google Scholar] [CrossRef]
- Shahid, M.; Mahmood, F.; Hussain, S.; Shahzad, T.; Haider, M.Z.; Noman, M.; Mushtaq, A.; Fatima, Q.; Ahmed, T.; Mustafa, G. Enzymatic detoxification of azo dyes by a multifarious Bacillus sp. strain MR-1/2-bearing plant growth-promoting characteristics. 3 Biotech 2018, 8, 425. [Google Scholar] [CrossRef]
- Vineh, M.B.; Saboury, A.A.; Poostchi, A.A.; Ghasemi, A. Biodegradation of phenol and dyes with horseradish peroxidase covalently immobilized on functionalized RGO-SiO2 nano composite. Int. J. Biol. Macromol. 2020, 164, 4403–4414. [Google Scholar] [CrossRef] [PubMed]
- Navas, L.E.; Carballo, R.; Levin, L.; Berretta, M.F. Fast decolorization of azo dyes in alkaline solutions by a thermostable metal-tolerant bacterial laccase and proposed degradation pathways. Extremophiles 2020, 24, 705–719. [Google Scholar] [CrossRef]
- Kalsoom, U.; Bhatti, H.N.; Aftab, K.; Amin, F.; Jesioncowski, T.; Bilal, M. Biocatalytic potential of Brassica oleracea L. var. botrytis leaves peroxidase for efficient degradation of textile dyes in aqueous medium. J. Bioprocess. Biosyst. Eng. 2023, 46, 453–465. [Google Scholar]
- Gao, Y.; Wang, M.; Shah, K.; Kalra, S.S.; Rome, L.H.; Mahendra, S. Decolourization and detoxification of synthetic dye compounds by laccase immobilized in vault nanoparticles. Bioresour. Technol. 2022, 351, 127040. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, T.; Kulla, H.G.; Leisinger, T. Properties of purified orange II azoreductase, the enzyme initiating azo dye degradation by Pseudomonas KF46. Eur. J. Biochem. 1982, 129, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Georgiou, D.; Hatiras, J.; Aivasidis, A. Microbial immobilization in a two-stage fixed-bed-reactor pilot plant for on-site anaerobic decolorization of textile wastewater. J. Enzym. Microb. Technol. 2005, 37, 597–605. [Google Scholar] [CrossRef]
- Jadhav, S.B.; Phugare, S.S.; Patil, P.S.; Jadhav, J.P. Biochemical degradation pathway of textile dye removal red and subsequent toxicological evaluation by cytotoxicity, genotoxicity and oxidative stress studies. Int. Biodeterior. Biodegrad. 2011, 65, 733–743. [Google Scholar] [CrossRef]
- Phugare, S.S.; Kalyani, D.C.; Surwase, S.N.; Jadhav, J.P. Ecofriendly degradation, decolorization and detoxification of textile effluent by a developed bacterial consortium. Ecotoxicol. Environ. Saf. 2011, 74, 1288–1296. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Zimmermann, W. Decolorization of industrial effluents containing reactive dyes by actinomycetes. FEMS Microbiol. Lett. 1993, 107, 157–161. [Google Scholar] [CrossRef] [PubMed]
- Saratale, R.G.; Saratale, G.D.; Cheng, J.S.; Govindwar, S.P. Bacterial decolorization and degradation of azo dyes. A review. J. Taiwan Inst. Chem. Eng. 2011, 42, 138–157. [Google Scholar] [CrossRef]
- Khan, R.; Bhawan, P.; Fulekar, M.H. Microbiol decolourization and degradation of synthetic dyes: A review. Rev. Environ. Sci. Bio/Technol. 2013, 12, 75–97. [Google Scholar] [CrossRef]
- Yang, H.Y.; Jia, R.B.; Chen, B.; Li, L. Degradation of recalcitrant aliphatic and aromatic hydrocarbons by a dioxin- degrader Rhodococcus sp. strain p52. Environ. Sci. Pollut. Res. 2014, 21, 11086–11093. [Google Scholar] [CrossRef]
- Nguyen, T.A.; Fu, C.C.; Juang, R.S. Biosorption and biodegradation of a sulphur dye in high-strength dyeing wastewater by Acidithiobacillus thiooxidans. J. Environ. Manag. 2016, 182, 265–271. [Google Scholar] [CrossRef]
- Louati, I.; Ellaumi-Mseddi, J.; Cheikhrouhou, W.; Hadrich, B.; Nasri, M.; Aifa, S.; Woodword, S.; Mechichi, T. Simultaneous cleanup of reactive black 5 and cadmium by a desert soil bacterium. J. Ecotoxicol. Environ. Saf. 2020, 190, 110103. [Google Scholar] [CrossRef] [PubMed]
- Montanez-Barragan, B.; Sanz-Martin, J.L.; Gutierrez-Macias, P.; Barragan-Huerta, B.E. Azo-dyes decolourization under high alkalinity and salinity conditions by Halomonas sp. in batch and packed bed reactor. J. Extrem. 2020, 20, 239–247. [Google Scholar]
- Shi, Y.; Yang, Z.; Xing, L.; Zhou, J.; Ren, J.; Ming, L.; Hun, Z.; Li, X.; Zhang, D. Ethanol as an efficient cosubstrate for biodegradation of azo dyes by Providencia rettgeri, mechanistic analysis based on kinetics, pathways and genomics. Bioresour. Technol. 2021, 31, 124117. [Google Scholar] [CrossRef] [PubMed]
- Fareed, A.; Zaffar, H.; Bilal, M.; Hussain, J.; Jackson, C.; Naqvi, T.A. Decolourization of azo-dyes by a novel aerobic bacterial strain Bacillus cereus strain ROC. PLoS ONE 2022, 17, e0269559. [Google Scholar] [CrossRef]
- Srinivasan, S.; Bankole, P.O.; Sadasivam, S.K. Biodecolorization and degradation of textile azo dyes using Lysinibacilllus sphaericus MTCC9523. Front. Environ. Sci. 2022, 10, 990855. [Google Scholar] [CrossRef]
- Du, L.; Wu, H.; Li, G.; Wei, Y.; Wang, F.; Xu, L.; Dong, X. Efficient degradation and decolorization of triphenyl methane dyes by Serratia sp. WKD under extreme environmental conditions and the mechanism. Int. Biodeterior. Biodegrad. 2023, 179, 105565. [Google Scholar]
- Joshi, A.; Hinsu, A.; Kothari, R. Evaluating the efficacy of bacterial consortium for decolourization of diazo dye mixture. J. Arch. Microbiol. 2022, 204, 515. [Google Scholar]
- Ayed, L.; Ladhari, N.; Achour, S.; Chaieb, K. Decolorization of reactive yellow 174 dye in real textile wastewater by active consortium; experimental factorial design for bioremediation process optimization. J. Text. Inst. 2020, 112, 1449–1459. [Google Scholar] [CrossRef]
- Guo, G.; Hao, J.; Tian, F.; Liu, C.; Ding, K.; Xu, J.; Zhou, W.; Guan, Z. Decolourization and detoxification of azo dyes by halo-alkaliphilic bacterium consortium: Systematic investigations of performance, pathway and metagenome. J. Ecotoxicol. Environ. Saf. 2020, 204, 111073. [Google Scholar] [CrossRef]
- Bera, S.P.; Shah, M.P.; Godhaniya, M. Microbial remediation of textile dye acid orange by a novel bacterial consortium SPB 92. Front. Environ. Sci. 2022, 10, 930616. [Google Scholar] [CrossRef]
- Neihsial, R.; Singha, N.A.; Singh, A.K. Taxonomic diversity and predictive metabolic functions of a heavy metal tolerant multiple azo dye degrading bacterial consortium from textile effluents. Int. Biodeterior. Biodegrad. 2022, 171, 105421. [Google Scholar] [CrossRef]
- Barathi, S.; Aruljothi, K.N.; Karthik, C.; Padikasan, I.A.; Ashokkumar, V. Biofilm mediated decolorization and degradation of reactive red 170 dye by the bacterial consortium isolated from the dyeing industry wastewater sediments. Chemosphere 2022, 286, 131914. [Google Scholar] [CrossRef] [PubMed]
- Franca, R.D.G.; Viera, A.; Corvalho, G.; Oehmen, A.; Pinheiro, H.M.; Barreto Crespo, M.T.; Lourence, N.D. Oerskovia paurometabola can efficiently decolourize azo dyes Acid Red 14 and remove its recalcitrant metabolite. J. Ecotoxicol. Environ. Saf. 2020, 191, 110007. [Google Scholar] [CrossRef] [PubMed]
- Saravanan, S.; Carolina C, F.; Kumar, P.S.; Chitra, B.; Rangasamy, G. Biodegradation of textile dye rhodamine-B by Brevundimonas diminuta and screening of their breakdown metabolites. Chemosphere 2022, 308, 136–266. [Google Scholar] [CrossRef]
- Gianolini, J.E.; Britas, C.N.; Mulreedy, C.B.; Trelles, J.A. Hyperstabilization of a thermophile bacterial laccase and it’s application for industrial dyes degradation. 3 Biotech 2020, 10, 208. [Google Scholar]
- Rajashekarappa, K.K.; Mahadevan, G.M.; Neelagund, S.E.; Madhuri Sathynarayana, M.; Vijaya, D.; Mulla, S.I. Decolorization of amaranth RI and fast red E azo dyes by thermophilic Geobacillus thermoleovorans KNG 112. J. Chem. Technol Biotech. 2022, 97, 482–489. [Google Scholar] [CrossRef]
- Ebihara, K.; Yoshikawa, J.; Horiguchi, H.; Amochi, S. Decolorization of cationic dyes under alkaline conditions by Iodidimonas sp. Q-1, multicopper oxidase. J. Biosci. Bioeng. 2022, 133, 323–328. [Google Scholar] [CrossRef]
- Dong, H.; Guo, T.; Zhang, W.; Ying, H.; Wang, P.; Wang, Y.; Chen, Y. Biochemical characterization of a novel azo-reductase from Streptomyces sp. application in eco-friendly decolorization of azo-dyes wastewater. J. Int. J. Biol. Macromol. 2019, 140, 1037–1046. [Google Scholar]
- Adenan, N.H.; Lim, Y.Y.; Ting, A.S.Y. Removal of triphenylmethane dyes by Streptomyces bacillaris: A study on decolorization, enzymatic reaction and toxicity of treated dye solution. J. Environ. Manag. 2022, 318, 115520. [Google Scholar] [CrossRef]
- Raja, M.M.M.; Raja, A.; Salique, S.M.; Gajalakshmi, P. Studies on effect of marine actinomycetes on amido black (azo dye) decolorization. J. Chem. Pharm. Res. 2016, 8, 640–644. [Google Scholar]
- Blánquez, A.; Rodríguez, J.; Brissos, V.; Mendes, S.; Martins, L.O.; Ball, A.S.; Arias, M.E.; Hernández, M. Decolorization and detoxification of textile dyes using a versatile Streptomyces laccase-natural mediator system. Saudi J. Biol. Sci. 2019, 26, 913–920. [Google Scholar] [CrossRef] [PubMed]
- Kameche, K.; Amrani, S.; Mouzaoui, S.; Aït-Amar, H. Biodegradation of diazo dye Evans blue by four strains of Streptomyces isolated from soils of Algeria. Biocatal. Agric. Biotechnol. 2022, 46, 102529. [Google Scholar] [CrossRef]
- Jamee, R.; Siddique, R. Biodegradation of synthetic dyes of textile effluent by micro-organisms: An environmentally and economically sustainable approach. Eur. J. Microb. Immunol. 2019, 9, 114–118. [Google Scholar] [CrossRef] [PubMed]
- Gangola, S.; Bhatt, P.; Chaudhary, P.; Khati, P.; Kumar, N.; Sharma, A. Bioremediation of industrial waste using microbial diversity. In Microbial Biotechnology in Environmental Monitoring and Cleanup; IGI Global: Hershey, PA, USA, 2018; pp. 1–27. [Google Scholar]
- Mahalakshmi, S.; Lakshmi, D.; Menaga, U. Biodegradation of different concentration of dye (congo red dye) by using green and blue-green algae. Int. Environ. Res. 2015, 9, 735–744. [Google Scholar]
- Mahajan, P.; Kaushal, J. Phytoremediation of azo dye methyl red by microalgal Chara vulgaris L.: Kinetics and equilibrium studies. Environ. Sci. Pollut. Res. 2020, 27, 26406–26418. [Google Scholar] [CrossRef]
- Boulkhessaim, S.; Gacem, A.; Khan, S.H.; Amari, A.; Yadav, V.K.; Harharah, H.N.; Elkhaleefa, A.M.; Yadav, K.K.; Rather, S.; Ahn, H.J.; et al. Emerging trends in the remediation of persistent organic pollutants using nanomaterials and related processes: A review. Nanomaterials 2022, 12, 2148. [Google Scholar] [CrossRef]
- Dellamatrice, P.M.; Silva-Stenico, M.E.; de Moraes, L.A.B.; Fiore, M.F.; Monteiro, R.T.S. Degradation of textile dyes by cyanobacteria. Braz. J. Microbiol. 2017, 48, 25–31. [Google Scholar] [CrossRef]
- Alprol, A.E.; Heneash, A.M.M.; Ashour, M.; Abulnaza, K.M.; Alhashmialmeer, D.; Mansour, A.T.; Sharawi, Z.Z.; Abu-Saled, M.A.; Abomohra, A.E.F. Potential applications of Arthrospira platensis lipid free biomass in bioremediation of organic dye from industrial textile effluents and its influence on marine rotifer (Brachionus plicatilis). Materials 2021, 16, 4446. [Google Scholar] [CrossRef]
- Mansour, A.T.; Alprol, A.E.; Abualnaja, K.M.; El-Beltagi, H.S.; Ramadan, K.M.A.; Ashour, M. The using of nano-particles of microalgae in remediation of toxic dye from industrial wastewater: Kinetic and isotherm studies. Materials 2022, 15, 3922. [Google Scholar] [CrossRef]
- Ngo, A.C.R.; Tischler, D. Microbial Degradation of azo dyes: Approaches and prospects for a hazard-free conversion by microorganisms. Int. J. Environ. Res. Public Health 2022, 19, 4740. [Google Scholar] [CrossRef]
- Jafari, N.; Soudib, M.R.; Kasra-Kermanshahi, R. Biodegradation perspectives of azo dyes by yeasts. Microbiology 2014, 83, 184–197. [Google Scholar] [CrossRef]
- Jadhav, S.U.; Kalme, S.D.; Govindwar, S.P. Biodegradation of Methyl red by Galactomyces geotrichum MTCC 1360. Int. Biodet. Biodeg. 2008, 62, 135–142. [Google Scholar] [CrossRef]
- Guo, G.; Tian, F.; Zhao, Y.; Tang, M.; Liu, W.; Liu, C.; Xue, S.; Kong, W.; Sun, Y.; Wang, S. Aerobic decolorization and detoxification of acid scarlet GR by a newly isolated salt-tolerant yeast strain Galactomyces geotrichum GG. Int. Biodeterior. Biodegrad. 2019, 145, 104818. [Google Scholar] [CrossRef]
- Ali, S.S.; Al-Tohamy, R.; Koupra, E.; EL-Naggar, A.H.; Kornaros, M.; Sun, J. Valorizing lignin-like dyes and textile dyeing wastewater by a newly constructed lipid producing and lignin modifying olraginous yeast consortioum valued for biodiesel and bioremediation. J. Hazard. Mater. 2021, 403, 123575. [Google Scholar] [CrossRef] [PubMed]
- Salt, D.E.; Smith, R.D.; Raskin, I. Phytoremediation. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998, 49, 643–668. [Google Scholar] [CrossRef] [PubMed]
- Greipsson, S. Phytoremediation. Nat. Educ. Knowl. 2011, 3, 7. [Google Scholar]
- Viana, D.G.; Filho, F.B.E.; Pires, F.R.; Soares, M.B.; Ferreira, A.D.; Bonomo, R.; Martins, L.F. In situ barium phytoremediation in flooded soil using Typha domingensis under different planting densities. Ecotoxicol. Environ. Saf. 2021, 210, 111890. [Google Scholar] [CrossRef]
- Wei, Z.; Van Le, Q.; Peng, W.; Yang, Y.; Yang, H.; Gu, H.; Lam, S.S.; Sonne, C. A review on phytoremediation of contaminants in air, water and soil. J. Hazard. Mater. 2021, 403, 123658. [Google Scholar] [CrossRef]
- Kafle, A.; Timilsina, A.; Gautam, A.; Adhikari, K.; Bhattarai, A.; Aryal, N. Phytoremediation: Mechanisms, plant selection and enhancement by natural and synthetic agents. Environ. Adv. 2022, 8, 100203. [Google Scholar] [CrossRef]
- Alsafran, M.; Usman, K.; Ahmed, B.; Rizwan, M.; Saleem, M.H.; Al Jabri, H. Understanding the phytoremediation mechanisms of potentially toxic elements: A proteomic overview of recent advances. Front. Plant Sci. 2022, 13, 881242. [Google Scholar] [CrossRef]
- Biju, L.M.; Gayathri, K.V.; Kumar, P.S.; Kavitha, R.; Rajagopal, R.; Ranasamy, G. Application of Salvinia species in remediation of reactive mix azo-dyes and Cr- its pathway eleucidation. Environ. Res. 2023, 216, 114635. [Google Scholar] [CrossRef] [PubMed]
- Rane, N.R.; Chandanshive, V.V.; Watharkar, A.D.; Khandare, R.V.; Patil, T.S.; Pawar, P.K.; Govindwar, S.P. Phytoremediation of sulfonated remazol red dye and textile effluents by Alternanthera philoxeroides: An anatomical, enzymatic and pilot scale study. Water Res. 2015, 83, 271–281. [Google Scholar] [CrossRef] [PubMed]
- Imron, M.F.; Kurniawan, S.B.; Soegianto, A.; Wahyudianto, F.E. Phytoremediation of methylene blue using duckweed (Lemna minor). Heliyon 2019, 5, e02206. [Google Scholar] [CrossRef]
- Baldawi, I.A.A.; Abdullah, S.R.S.; Anuar, N.; Hasan, H.A. Phytotransformation of methylene blue from water using aquatic plant (Azolla pinnata). Environ. Technol. Innov. 2018, 11, 15–22. [Google Scholar] [CrossRef]
- Mishra, B.; Varjani, S.; Pradhan, I.; Ekambaram, N.; Teixeira, J.A.; Ngo, H.H.; Guo, W. Insights into inter-disciplinary approaches for bioremediation of organic pollutants: Innovations, challenges and perspectives. Proc. Natl. Acad. Sci. India B Biol. Sci. 2020, 90, 951–958. [Google Scholar] [CrossRef]
- Liu, L.; Bilal, M.; Duan, X.; Iqbal, H.M. Mitigation of environmental pollution by genetically engineered bacteria- current challenges and future perspectives. Sci. Total Environ. 2019, 667, 444–454. [Google Scholar] [CrossRef] [PubMed]
- Jafari, M.; Danesh, Y.R.; Goltapeh, E.M.; Varma, A. Bioremediation and genetically modified organisms. In Fungi as Bioremediators; Springer: Berlin/Heidelberg, Germany, 2013; pp. 433–451. [Google Scholar]
- Sanghvi, G.; Thanki, A.; Pandey, S.; Singh, N.K. Engineered bacteria for bioremediation. In Bioremediation of Pollutants; Elsevier: Amsterdam, The Netherlands, 2020; pp. 359–374. [Google Scholar]
- Bu, T.; Yang, R.; Zheng, Y.; Cai, Y.; Tang, Z.; Li, C.; Wu, Q.; Chen, H. Improving decolorization of dyes by laccase from Bacillus licheniformis by random and site directed mutagenesis. PeerJ 2020, 8, e10267. [Google Scholar] [CrossRef] [PubMed]
- Dixit, S.; Garg, S. Biodegradation of environmentally hazardous azo-dyes and aromatic amines using Klebsiella pneumoniae. J. Environ. Eng. 2018, 144, 04018035. [Google Scholar] [CrossRef]
- Chang, J.S.; Kuo, T.S.; Chao, V.P.; Ho, J.Y.; Lin, P.J. Azo dye decolorization with a mutant E. coli strain. Biotechnol. Lett. 2000, 22, 807–812. [Google Scholar] [CrossRef]
- Fomina, M.; Gadd, G.M. Biosorption current perspectives can concept, definition and applications. Bioresour. Technol. 2014, 160, 3–14. [Google Scholar] [CrossRef]
- Kour, D.; Kaur, T.; Devi, R.; Yadav, A.; Singh, M.; Joshi, D. Beneficial mirobiomes for bioremediation of diverse contaminated environments for environmental sustainability: Present status and future challenges. Environ. Sci. Pollut. Res. 2021, 28, 24917–24939. [Google Scholar] [CrossRef] [PubMed]
- Bayat, M.; Salehi, E.; Mahdieh, M. Chromochloris zofingiensis microalgae as a potential dye absorbent; adsorption thermo-kinetic, isothermal and process optimization. Algal Res. 2023, 71, 103043. [Google Scholar] [CrossRef]
- Sun, W.; Sun, W.; Wang, Y. Biosorption of direct fast scarlet 4BS from aqueous solution using the green tide causing marine algae Enteromorpha prolifera. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2019, 223, 117347. [Google Scholar] [CrossRef] [PubMed]
- Silva, F.; Nascimento, L.; Brito, M.; Silva, K.; Paschoal, W., Jr.; Fujiyama, R. Biosorption of methylene blue dye using natural biosorbents made from weeds. Materials 2019, 12, 2486. [Google Scholar] [CrossRef]
- Flores Alarcon, M.A.D.; Pacheco, C.R.; Bustos, K.G.; Meza, K.T.; Teran, H.F.; Tanaka, D.A.P.; Andrade, G.I.C.; Teran, H.R. Efficient dye removal from real textile wastewater using orange seed powder as suitable bioadsorbent and membrane technology. Water 2022, 14, 4104. [Google Scholar] [CrossRef]
- Elmoutez, S.; Abushaban, A.; Necibi, M.C.; Sillanpaa, M.; Liu, J.; Dhiba, D.; Chehbouni, A.; Taky, M. Design and operational aspects of anaerobic bioreactor for efficient wastewater treatment and biogas production. J. Environ. Chall. 2023, 10, 100671. [Google Scholar] [CrossRef]
- Manzoor, K.; Khan, S.J.; Yasmeen, M.; Jamal, Y.; Arshad, M. Assessment of anaerobic membrane distillation bioreactor hybrid system at mesophilic and thermophilic temperature treating textile wastewater. J. Water Process Eng. 2022, 46, 102603. [Google Scholar] [CrossRef]
- Jin, Y.; Wang, D.; Zhang, W. Newly designed hydrolysis acidification flat ceramic membrane bioreactor for treating high-strength dyeing wastewater. Int. J. Environ. Res. Public Health 2019, 16, 777. [Google Scholar] [CrossRef]
- Castro, F.D.; Bassin, J.P.; Alves, P.L.M.; Sant’Anna, G.L.; Dezotti, M. Reactive orange 16 dye degradation in anaerobic and aerobic MBBR coupled with ozonation; addressing pathways and performance. Int. J. Environ. Sci. Technol. 2021, 18, 1991–2010. [Google Scholar] [CrossRef]
- Zhang, W.; Liu, F.; Wang, D.; Jin, Y. Impact of reactor configuration on treatment performance and microbial diversity in treating high- strength dyeing wastewater. anaerobic flat- sheet ceramic membrane bioreactor versus upflow anaerobic sludge blanket reactor. Bioresour. Technol. 2018, 269, 269–275. [Google Scholar]
- Balrabe, Y.B.; Oumarou, I.N.M.; Korony, S.A.; Adjama, I.; Baraze, I.R.A. Photooxidation of organic dye by Fe2O3 nanoparticles; catalyst, electron acceptor and polyurethane membrane effects. J. Nanotechnol. 2023, 2023, 1292762. [Google Scholar]
- Garg, D.; Sridhar, K.; Stephen Inbaraj, B.; Chawla, P.; Tripathi, M.; Sharma, M. Nano-biofertilizer formulations for agriculture: A systematic review on recent advances and prospective applications. Bioengineering 2023, 10, 1010. [Google Scholar] [CrossRef] [PubMed]
- Bala, S.; Sharma, M.; Dashora, K.; Siddiqui, S.; Diwan, D.; Tripathi, M. Nanomaterials based sustainable bioenergy production systems: Current trends and future prospects. Nanofabrication 2022, 7, 314–324. [Google Scholar] [CrossRef]
- Darwesh, O.M.; Li, H.; Matter, I.A. Nano-bioremediation of textile industry wastewater using immobilized CuO-NPs myco-synthesized by a novel Cu-resistant Fusarium oxysporum OSF18. Environ. Sci. Pollut. Res. 2023, 30, 16694–16706. [Google Scholar] [CrossRef] [PubMed]
- Rani, M.; Shanker, U. Removal of organic dyes by functionalized nanomaterials. In Handbook of Green and Sustainable Nanotechnology; Shanker, U., Hussain, C.M., Rani, M., Eds.; Springer: Cham, Switzerland, 2023; pp. 1267–1298. [Google Scholar] [CrossRef]
- Sun, L.; Mo, Y.; Zhang, L. A mini review on bio-electrochemical systems for the treatment of azo-dye wastewater: State-of-the-art and future prospects. Chemosphere 2022, 294, 133801. [Google Scholar] [CrossRef] [PubMed]
- Das, A. Dye degradation using microbial fuel cell: A critical review. Int. J. Res. Appl. Sci. Eng. Technol. 2022, 10, 1745–1754. [Google Scholar] [CrossRef]
- Jain, A.; Zhen, H. Cathode-enhanced wastewater treatment in bio electro chemical system. Clean Water 2018, 1, 23. [Google Scholar] [CrossRef]
- Yang, M.; Korneel, R.; Reni, A.; Rozendal, R.A.; Yuan, Z.; Keller, J. Decolorization of azo dyes in bioelecrochemical systems. Environ. Sci. Technol. 2009, 43, 5137–5143. [Google Scholar]
- Yadav, A.; Kumar, P.; Rawat, D.; Gara, S.; Mukherjee, P.; Farooqi, F.; Roy, A.; Sundaram, S.; Sharma, R.S.; Mishra, V. Microbial fuel cells for mineralization and decolorization of azo dyes; recent advances in design and materials. Sci. Total Environ. 2022, 826, 154038. [Google Scholar] [CrossRef]
- Yaqoob, A.A.; Khatoon, A.; Mohd Setapar, S.H.; Umar, K.; Parveen, T.; Mohamad Ibrahim, M.N.; Ahmad, A.; Rafatullah, M. Outlook on the role of microbial fuel cells in remediation of environmental pollutants with electricity generation. Catalysts 2020, 10, 819. [Google Scholar] [CrossRef]
- Srivastava, A.; Rani, R.M.; Patle, D.S.; Kumar, S. Emerging bioremediation technologies for the treatment of textile wastewater containing synthetic dyes: A comprehensive review. J. Chem. Technol. Biotechnol. 2022, 97, 26–41. [Google Scholar] [CrossRef]
- Singh, S.; Kumar, V.; Datta, S.; Dhanjal, D.S.; Sharma, K.; Samuel, J.; Singh, J. Current advancement and future prospect of biosorbents for bioremediation. Sci. Total Environ. 2020, 709, 135895. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.; Kumari, S.; Rath, S.; Das, S. Prospects and scope of microbial bioremediation for the restoration of the contaminated sites. In Microbial Biodegradation and Bioremediation; Elsevier: Amsterdam, The Netherlands, 2022; pp. 3–31. [Google Scholar]
- Shah, M.; Ahmed, S. Conventional strategies of bioremediation versus green nanoremediation. In Green Nanoremediation: Sustainable Management of Environmental Pollution; Springer International Publishing: Cham, Switzerland, 2023; pp. 333–356. [Google Scholar]
- Reddy, S.; Osborne, J. An insight on the advancements of biological technologies in the bioremediation of textile effluents. Urban Water J. 2022, 19, 468–480. [Google Scholar] [CrossRef]
- Yogalakshmi, K.N.; Das, A.; Rani, G.; Jaswal, V.; Randhawa, J.S. Nano-bioremediation: A new age technology for the treatment of dyes in textile effluents. Bioremediat. Ind. Waste Environ. Saf. Ind. Waste Its Manag. 2020, 1, 313–347. [Google Scholar]
- Ihsanullah, I.; Jamal, A.; Ilyas, M.; Zubair, M.; Khan, G.; Atieh, M.A. Bioremediation of dyes: Current status and prospects. J. Water Process Eng. 2020, 38, 101680. [Google Scholar] [CrossRef]
- Sudarshan, S.; Harikrishnan, S.; RathiBhuvaneswari, G.; Alamelu, V.; Aanand, S.; Rajasekar, A.; Govarthanan, M. Impact of textile dyes on human health and bioremediation of textile industry effluent using microorganisms: Current status and future prospects. J. Appl. Microbiol. 2023, 134, lxac064. [Google Scholar] [CrossRef]
- Nachiyar, C.V.; Rakshi, A.D.; Sandhya, S.; Jebasta, N.B.D.; Nellore, J. Developments in treatment technologies of dye-containing effluent: A review. Stud. Chem. Case Environ. Eng. 2023, 7, 100339. [Google Scholar] [CrossRef]
- Nishshanka, G.K.S.H.; Thevarajah, B.; Nimarshana, P.H.V.; Prajapati, S.K.; Ariyadasa, T.U. Real-time integration of microalgae-based bioremediation in conventional wastewater treatment plants: Current status and prospects. J. Water Process Eng. 2023, 56, 104248. [Google Scholar] [CrossRef]






| Bacteria | Dye | % Decolorization | Reference |
|---|---|---|---|
| Oerskovia paurometabola | Acid red 14 | 91% | [114] |
| Brevundimonas diminuta | Rhodamine–B | 90–95% | [115] |
| Gerobacillus stereothermophilus ATCC 10149 | Remazol brilliant blue-R | 90% | [116] |
| Gerobacillus thermoleovorans | Amaranth RI Fast red E | 99% | [117] |
| Serratia spp. | Malachite green Crystal violet | 96.5% | [107] |
| Iodidimonas spp. | Cationic dyes (Malachite green, crystal violet, methylene blue) | >90% | [118] |
| Lysinibacillus sphaericus | Reactive Yellow F3R Joyfix Red RB | 96.30% 92.71% | [106] |
| S. No. | Factors | Advanced Oxidation Processes | Biological Treatment | Phytoremediation | Nanotechnology | Biotechnological Approaches |
|---|---|---|---|---|---|---|
| 1. | Mechanism | Utilizes chemical oxidation to break down dyes and contaminants through highly reactive species (e.g., hydroxyl radicals). | Microorganisms, enzymes, and natural processes degrade dyes biologically. | Involves the use of plants to uptake, metabolize, or sequester dyes from the environment. | Utilizes nanomaterials or nanoparticles to adsorb, degrade, or facilitate the removal of dyes. | Relies on genetically modified or engineered microorganisms for enhanced dye degradation. |
| 2. | Speed of Treatment | Generally rapid and efficient in dye degradation. | Biodegradation rates can vary and may be slower than chemical oxidation. | Treatment rates can be relatively slow, influenced by plant growth and environmental conditions. | Offers fast and efficient removal through high surface area and reactivity. | Can be engineered for rapid and targeted dye degradation. |
| 3. | Selectivity | May not exhibit high selectivity and can degrade a wide range of dyes. | Microbes may exhibit selectivity towards specific dyes, affecting treatment effectiveness. | Plant species selection influences specificity, with variations in the range of dyes targeted. | Can be designed for selectivity through nanomaterial selection and modification. | Selectivity can be engineered by designing microorganisms with specific dye-degrading enzymes. |
| 4. | Environmental Impact | May generate secondary byproducts and require careful management. | Generally environmentally friendly, with lower chemical use and reduced environmental impact. | Environmentally friendly, as it relies on natural processes and plant uptake. | Impact depends on nanomaterials used, with potential environmental concerns. | Environmental impact may vary depending on genetic modifications, but aims for minimal harm. |
| 5. | Energy Requirements | Requires energy for chemical processes, potentially energy-intensive. | Typically energy-efficient, relying on microbial metabolism or natural processes. | Minimal energy requirements, as it mainly depends on plant growth. | Energy-efficient, but energy may be required for nanoparticle synthesis. | Energy-efficient, but genetic engineering may involve energy-intensive processes. |
| 6. | Scale-Up Complexity | Scaling up AOPs can be complex and may require advanced infrastructure. | Scaling biological treatment systems is generally feasible but can be size-dependent. | Scalability can be challenging for large-scale phytoremediation projects due to space and time requirements. | Nanotechnology can be scaled up relatively easily but requires careful engineering. | Scalability depends on the cultivation and maintenance of engineered microorganisms, which can be challenging. |
| 7. | Cost Effectiveness | Initial setup costs can be high due to equipment and chemical requirements. | Generally cost-effective in the long run due to lower operating costs and sustainability. | Cost-effectiveness depends on factors like plant species, maintenance, and project size. | Costs may vary depending on the nanomaterials used and their availability. | Costs can be higher due to research and development, genetic modification, and monitoring. |
| 8. | Regulatory Considerations | May face regulatory scrutiny due to chemical usage and potential byproduct generation. | Typically meets regulatory compliance easily, especially for non-genetically modified organisms. | Regulatory approval depends on plant species used and potential ecological impacts. | Regulatory concerns related to nanoparticle release and toxicity may apply. | Requires regulatory approval for genetically modified organisms and potential ecological impacts. |
| S. No. | Aspects | Conventional Techniques | Advanced Techniques |
|---|---|---|---|
| 1 | Treatment Principle | Relies mainly on natural processes and microbial action. | Integrates innovative approaches and advanced materials. |
| 2 | Effectiveness | May be limited in the removal of complex and recalcitrant dyes. | Generally, more effective in breaking down a wide range of dyes. |
| 3 | Speed of Treatment | Biodegradation rates can be slow. | Often faster due to enhanced microbial activity and optimized conditions. |
| 4 | Microbial Strains | Utilizes naturally occurring microorganisms. | May involve the use of genetically modified or engineered microorganisms. |
| 5 | Nutrient Requirements | Requires standard nutrients and conditions for microbial growth. | May require tailored nutrient supplementation for specific dye degradation. |
| 6 | pH and Temperature Control | Typically relies on ambient conditions. | Requires precise control of pH and temperature for optimal performance. |
| 7 | Dye Specificity | Some microbes may exhibit selectivity towards certain dyes. | Can target a broader range of dyes through microbial diversity or modifications. |
| 8 | Toxic Byproducts | May produce secondary pollutants or byproducts. | Tends to generate fewer toxic byproducts due to targeted degradation. |
| 9 | Resilience to Shock Loads | Vulnerable to shock loads and fluctuations in dye concentrations. | Better equipped to handle variations in dye concentrations. |
| 10 | Scale-up Challenges | Scaling up conventional bioremediation processes can be challenging. | Advanced techniques may offer more scalability and adaptability. |
| 11 | Sustainability | Moderately sustainable, but environmental impacts may vary. | Aims for increased sustainability through optimized processes. |
| 12 | Costs | Typically lower initial costs but may require longer treatment times. | May have higher initial setup costs but can be more cost-effective in the long term. |
| 13 | Regulatory Compliance | Conventional methods may require fewer regulatory approvals. | Advanced techniques may face additional regulatory scrutiny due to genetic modifications or novel materials. |
| 14 | Treatment Principle | Relies mainly on natural processes and microbial action. | Integrates innovative approaches and advanced materials. |
| 15 | Effectiveness | May be limited in the removal of complex and recalcitrant dyes. | Generally, more effective in breaking down a wide range of dyes. |
| 16 | Speed of Treatment | Biodegradation rates can be slow. | Often faster due to enhanced microbial activity and optimized conditions. |
| 17 | Selectivity | Limited selectivity and may not effectively target specific dyes | Can be engineered for selectivity targeting specific dye pollutants |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tripathi, M.; Singh, S.; Pathak, S.; Kasaudhan, J.; Mishra, A.; Bala, S.; Garg, D.; Singh, R.; Singh, P.; Singh, P.K.; et al. Recent Strategies for the Remediation of Textile Dyes from Wastewater: A Systematic Review. Toxics 2023, 11, 940. https://0-doi-org.brum.beds.ac.uk/10.3390/toxics11110940
Tripathi M, Singh S, Pathak S, Kasaudhan J, Mishra A, Bala S, Garg D, Singh R, Singh P, Singh PK, et al. Recent Strategies for the Remediation of Textile Dyes from Wastewater: A Systematic Review. Toxics. 2023; 11(11):940. https://0-doi-org.brum.beds.ac.uk/10.3390/toxics11110940
Chicago/Turabian StyleTripathi, Manikant, Sakshi Singh, Sukriti Pathak, Jahnvi Kasaudhan, Aditi Mishra, Saroj Bala, Diksha Garg, Ranjan Singh, Pankaj Singh, Pradeep Kumar Singh, and et al. 2023. "Recent Strategies for the Remediation of Textile Dyes from Wastewater: A Systematic Review" Toxics 11, no. 11: 940. https://0-doi-org.brum.beds.ac.uk/10.3390/toxics11110940