Seed Train Optimization in Microcarrier-Based Cell Culture Post In Situ Cell Detachment through Scale-Down Hybrid Modeling
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Line and Culture Medium
2.2. Subculture in T-Flask
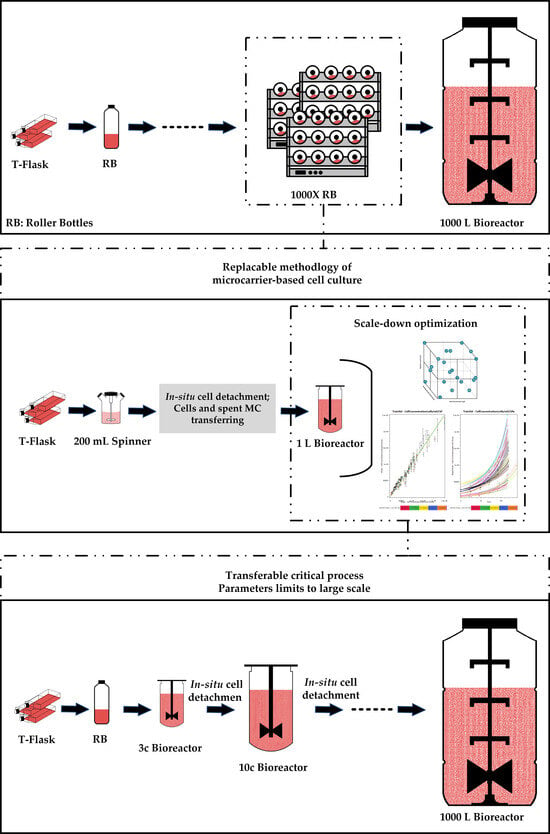
2.3. Subculture on MCs by Consecutive Cell Detachment–Attachment without Cell and MC Separation
2.4. Subculture on MCs by Cell and MC Separation after In Situ Cell Detachment
2.5. Repetitive Recolonization of the Spent MC
2.6. MC Culture in the Bioreactor for the DoE Study
2.7. Subculturing in Two Consecutive Bioreactors
2.8. Minimum Agitation Speed Determination
2.9. Sampling and Analytical Methods
2.10. Hybrid Modeling of Cellular Growth on MC after In Situ Cell Detachment
2.10.1. Parameter Selection for the DoE Study
- = mean specific energy dissipation rate (m2/s3)
- ν = kinematic viscosity (cm2/s)
- P = power consumption (kg.m2/s3)
- = density (g/cm3)
- V = working volume of the reactor (cm3)
- Np = power number of the bioreactor
- N = agitation speed (1/s)
- Di = impeller diameter.
| Agitation Speed (rpm) | Tip Speed (m/s) | Re | (m2/s3) | λ (μm) |
|---|---|---|---|---|
| 70 | 0.183 | 2607 | 0.000106 | 190.50 |
| 100 | 0.262 | 3724 | 0.0031 | 145.79 |
| 130 | 0.340 | 4841 | 0.0068 | 119.75 |
2.10.2. DoE Study
2.10.3. Hybrid Modeling
3. Results
3.1. Subculture on MCs by Consecutive Cell Detachment–Attachment without Cell and MC Separation
3.2. Subculture on MCs by Cell and MC Separation after In Situ Cell Detachment
3.3. Repetitive Recolonization of the Spent MC
3.4. Hybrid Modeling of Cellular Growth on MC after In Situ Cell Detachment
3.5. Validation of Predicted Culture Conditions in Two Consecutive MC Cultures in a Bioreactor
4. Discussion
4.1. Subculture on MCs by Consecutive Cell Detachment–Attachment without Cell and MC Separation
4.2. Subculture on MCs by Cell and MC Separation after In Situ Cell Detachment
4.3. Repetitive Recolonization of Spent MC
4.4. DoE Study and Hybrid Process Modeling Outcome
- H = height at working volume (cm)
- DT = tank diameter (cm)
- Wi = impeller blade width (cm).
4.5. Scale-Up Outlook
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ellebedy, A.H.; Ahmed, R. Antiviral Vaccines: Challenges and Advances. In The Vaccine Book; Elsevier: Amsterdam, The Netherlands, 2016; pp. 283–310. [Google Scholar] [CrossRef]
- Hajiaghapour Asr, M.; Dayani, F.; Saedi Segherloo, F.; Kamedi, A.; Neill, A.O.; MacLoughlin, R.; Doroudian, M. Lipid Nanoparticles as Promising Carriers for mRNA Vaccines for Viral Lung Infections. Pharmaceutics 2023, 15, 1127. [Google Scholar] [CrossRef]
- Liu, T.; Liang, Y.; Huang, L. Development and Delivery Systems of mRNA Vaccines. Front. Bioeng. Biotechnol. 2021, 9, 718753. [Google Scholar] [CrossRef]
- Zhang, J.; Qiu, Z.; Wang, S.; Liu, Z.; Qiao, Z.; Wang, J.; Duan, K.; Nian, X.; Ma, Z.; Yang, X. Suspended cell lines for inactivated virus vaccine production. Expert Rev. Vaccines 2023, 22, 468–480. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, L.; Ye, Z.; Li, L.; Yang, L.; Gong, W. Next-Generation TB Vaccines: Progress, Challenges, and Prospects. Vaccines 2023, 11, 1304. [Google Scholar] [CrossRef] [PubMed]
- Gallo-Ramírez, L.E.; Nikolay, A.; Genzel, Y.; Reichl, U. Bioreactor concepts for cell culture-based viral vaccine production. Expert Rev. Vaccines 2015, 14, 1181–1195. [Google Scholar] [CrossRef]
- Kiesslich, S.; Kamen, A.A. Vero cell upstream bioprocess development for the production of viral vectors and vaccines. Biotechnol. Adv. 2020, 44, 107608. [Google Scholar] [CrossRef]
- Chou, A.-H.; Liu, C.-C.; Chang, C.-P.; Guo, M.-S.; Hsieh, S.-Y.; Yang, W.-H.; Chao, H.-J.; Wu, C.-L.; Huang, J.-L.; Lee, M.-S.; et al. Pilot scale production of highly efficacious and stable enterovirus 71 vaccine candidates. PLoS ONE 2012, 7, e34834. [Google Scholar] [CrossRef] [PubMed]
- Aubrit, F.; Perugi, F.; Léon, A.; Guéhenneux, F.; Champion-Arnaud, P.; Lahmar, M.; Schwamborn, K. Cell substrates for the production of viral vaccines. Vaccine 2015, 33, 5905–5912. [Google Scholar] [CrossRef] [PubMed]
- European Medicine Agency. Assessment Report Ervebo. Available online: https://www.ema.europa.eu/en/documents/assessment-report/ervebo-epar-public-assessment-report_en.pdf (accessed on 17 October 2019).
- Berry, E.J. Commercial scale process for production of PRRSV. U.S. Patent 9944902B2, 17 April 2018. [Google Scholar]
- Trabelsi, K.; Zakour, M.B.; Jordan, I.; Sandig, V.; Rourou, S.; Kallel, H. Development of an efficient veterinary rabies vaccine production process in the avian suspension cell line AGE1.CR.pIX. BMC Biotechnol. 2022, 22, 17. [Google Scholar] [CrossRef]
- van Wezel, A.L. Growth of cell-strains and primary cells on micro-carriers in homogeneous culture. Nature 1967, 216, 64–65. [Google Scholar] [CrossRef]
- Yang, J.; Guertin, P.; Jia, G.; Lv, Z.; Yang, H.; Ju, D. Large-scale microcarrier culture of HEK293T cells and Vero cells in single-use bioreactors. AMB Express 2019, 9, 70. [Google Scholar] [CrossRef]
- Badenes, S.M.; Fernandes-Platzgummer, A.; Rodrigues, C.; Diogo, M.M.; Da Silva, C.L.; Cabral, J. Microcarrier Culture Systems for Stem Cell Manufacturing. Stem Cell Manufacturing; Elsevier: Amsterdam, The Netherlands, 2016; pp. 77–104. ISBN 9780444632654. [Google Scholar]
- Tsai, A.-C.; Pacak, C.A. Bioprocessing of Human Mesenchymal Stem Cells: From Planar Culture to Microcarrier-Based Bioreactors. Bioengineering 2021, 8, 96. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-Y.; Lin, Y.-W.; Kuo, C.-H.; Liu, W.-H.; Tai, H.-F.; Pan, C.-H.; Chen, Y.-T.; Hsiao, P.-W.; Chan, C.-H.; Chang, C.-C.; et al. Inactivated Enterovirus 71 Vaccine Produced by 200-L Scale Serum-Free Microcarrier Bioreactor System Provides Cross-Protective Efficacy in Human SCARB2 Transgenic Mouse. PLoS ONE 2015, 10, e0136420. [Google Scholar] [CrossRef] [PubMed]
- Derakhti, S.; Safiabadi-Tali, S.H.; Amoabediny, G.; Sheikhpour, M. Attachment and detachment strategies in microcarrier-based cell culture technology: A comprehensive review. Mater. Sci. Eng. C Mater. Biol. Appl. 2019, 103, 109782. [Google Scholar] [CrossRef] [PubMed]
- El-Bagoury, G.F. Optimizing Culture Conditions for Increasing Production of Vero Cell. Available online: https://bvmj.journals.ekb.eg/article_123444_12db61f5cdd6ea0d45476dbc8b709b1a.pdf (accessed on 1 March 2019).
- Sun, X.; Zhang, Y.; Tan, W.; Zhou, Y.; Hua, P. Attachment kinetics of Vero cells onto CT-3 microcarriers. J. Biosci. Bioeng. 2000, 90, 32–36. [Google Scholar] [CrossRef] [PubMed]
- Souza, M.C.d.O.; Da Freire, M.S.; Castilho, L.d.R. Influence of culture conditions on Vero cell propagation on non-porous microcarriers. Braz. Arch. Biol. Technol. 2005, 48, 71–77. [Google Scholar] [CrossRef]
- Cytiva, Application Note 29-0435-48 AB. Scale-Up of Adherent Vero Cells Grown on Cytodex Microcarriers Using Single-Use Bioprocessing Equipment. Available online: https://cdn.cytivalifesciences.com/api/public/content/digi-16539-pdf (accessed on 1 April 2014).
- Nienow, A.W.; Hewitt, C.J.; Heathman, T.R.; Glyn, V.A.; Fonte, G.N.; Hanga, M.P.; Coopman, K.; Rafiq, Q.A. Agitation conditions for the culture and detachment of hMSCs from microcarriers in multiple bioreactor platforms. Biochem. Eng. J. 2016, 108, 24–29. [Google Scholar] [CrossRef]
- Sousa, M.; Fenge, C.; Rupprecht, J.; Tappe, A.; Greller, G.; Alves, P.; Carrondo, M.; Roldão, A. Process intensification for Peste des Petites Ruminants Virus vaccine production. Vaccine 2019, 37, 7041–7051. [Google Scholar] [CrossRef]
- Sun, X.; Zhang, Y. A cell-detaching reactor for inoculation of anchorage-dependent CHO and Vero cells between stepwise-expanded bioreactors. Biotechnol. Lett. 2007, 29, 697–701. [Google Scholar] [CrossRef]
- Kistner, O.; Barrett, P.N.; Mundt, W.; Reiter, M.; Schober-Bendixen, S.; Dorner, F. Development of a mammalian cell (Vero) derived candidate influenza virus vaccine. Vaccine 1998, 16, 960–968. [Google Scholar] [CrossRef]
- Sugawara, K.; Nishiyama, K.; Ishikawa, Y.; Abe, M.; Sonoda, K.; Komatsu, K.; Horikawa, Y.; Takeda, K.; Honda, T.; Kuzuhara, S.; et al. Development of Vero cell-derived inactivated Japanese encephalitis vaccine. Biologicals 2002, 30, 303–314. [Google Scholar] [CrossRef]
- Wu, W.; Orr-Burks, N.; Karpilow, J.; Tripp, R.A. Development of improved vaccine cell lines against rotavirus. Sci. Data 2017, 4, 170021. [Google Scholar] [CrossRef]
- Rai, A.; Pruitt, S.; Ramirez-Medina, E.; Vuono, E.A.; Silva, E.; Velazquez-Salinas, L.; Carrillo, C.; Borca, M.V.; Gladue, D.P. Identification of a Continuously Stable and Commercially Available Cell Line for the Identification of Infectious African Swine Fever Virus in Clinical Samples. Viruses 2020, 12, 820. [Google Scholar] [CrossRef]
- Ward, R.L.; Knowlton, D.R.; Pierce, M.J. Efficiency of human rotavirus propagation in cell culture. J. Clin. Microbiol. 1984, 19, 748–753. [Google Scholar] [CrossRef]
- Tsunemitsu, H.; Saif, L.J.; Jiang, B.M.; Shimizu, M.; Hiro, M.; Yamaguchi, H.; Ishiyama, T.; Hirai, T. Isolation, characterization, and serial propagation of a bovine group C rotavirus in a monkey kidney cell line (MA104). J. Clin. Microbiol. 1991, 29, 2609–2613. [Google Scholar] [CrossRef]
- Heathman, T.R.; Nienow, A.W.; Rafiq, Q.A.; Coopman, K.; Kara, B.; Hewitt, C.J. Agitation and aeration of stirred-bioreactors for the microcarrier culture of human mesenchymal stem cells and potential implications for large-scale bioprocess development. Biochem. Eng. J. 2018, 136, 9–17. [Google Scholar] [CrossRef]
- Nienow, A.W. Reactor engineering in large scale animal cell culture. Cytotechnology 2006, 50, 9–33. [Google Scholar] [CrossRef] [PubMed]
- Rafiq, Q.A.; Hanga, M.P.; Heathman, T.R.J.; Coopman, K.; Nienow, A.W.; Williams, D.J.; Hewitt, C.J. Process development of human multipotent stromal cell microcarrier culture using an automated high-throughput microbioreactor. Biotechnol. Bioeng. 2017, 114, 2253–2266. [Google Scholar] [CrossRef] [PubMed]
- Hewitt, C.J.; Lee, K.; Nienow, A.W.; Thomas, R.J.; Smith, M.; Thomas, C.R. Expansion of human mesenchymal stem cells on microcarriers. Biotechnol. Lett. 2011, 33, 2325–2335. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.; Giroux, D.; Hashimura, Y.; Wesselschmidt, R. Scale-down model screening study of key process parameters for anchorage-dependent stem cells using vertical-wheel bioreactors. Cytotherapy 2015, 17, S30. [Google Scholar] [CrossRef]
- Croughan, M.S.; Hamel, J.-F.; Wang, D.I.C. Hydrodynamic effects on animal cells grown in microcarrier cultures. 1987. Biotechnol. Bioeng. 2006, 95, 295–305. [Google Scholar] [CrossRef]
- Glaser, R.; Greenlea, Z.; Sha, M. Stimulating Growth. Cultivating Solutions: Power Number for Cell Culture Glass Vessels; Eppendorf: Hamburg, Germany, 2020. [Google Scholar]
- Bayer, B.; Duerkop, M.; Pörtner, R.; Möller, J. Comparison of mechanistic and hybrid modeling approaches for characterization of a CHO cultivation process: Requirements, pitfalls and solution paths. Biotechnol. J. 2023, 18, e2200381. [Google Scholar] [CrossRef]
- Nold, V.; Junghans, L.; Bayer, B.; Bisgen, L.; Duerkop, M.; Drerup, R.; Presser, B.; Schwab, T.; Bluhmki, E.; Wieschalka, S.; et al. Boost dynamic protocols for producing mammalian biopharmaceuticals with intensified DoE—A practical guide to analyses with OLS and hybrid modeling. Front. Chem. Eng. 2023, 4, 1044245. [Google Scholar] [CrossRef]
- Bayer, B.; Striedner, G.; Duerkop, M. Hybrid Modeling and Intensified DoE: An Approach to Accelerate Upstream Process Characterization. Biotechnol. J. 2020, 15, e2000121. [Google Scholar] [CrossRef]
- Luo, F.; Sun, H.; Geng, T.; Qi, N. Application of Taguchi’s method in the optimization of bridging efficiency between confluent and fresh microcarriers in bead-to-bead transfer of Vero cells. Biotechnol. Lett. 2008, 30, 645–649. [Google Scholar] [CrossRef]
- Takahashi, I.; Sato, K.; Mera, H.; Wakitani, S.; Takagi, M. Effects of agitation rate on aggregation during beads-to-beads subcultivation of microcarrier culture of human mesenchymal stem cells. Cytotechnology 2017, 69, 503–509. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ouyang, F. Bead-to-bead transfer of Vero cells in microcarrier culture. Cytotechnology 1999, 31, 221–224. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Sato, Y.; Tada, Y.; Suzuki, Y.; Takahashi, R.; Okanojo, M.; Nakashima, K. Facile bead-to-bead cell-transfer method for serial subculture and large-scale expansion of human mesenchymal stem cells in bioreactors. Stem Cells Transl. Med. 2021, 10, 1329–1342. [Google Scholar] [CrossRef] [PubMed]
- Souza, M.C.O.; Freire, M.S.; Schulze, E.A.; Gaspar, L.P.; Castilho, L.R. Production of yellow fever virus in microcarrier-based Vero cell cultures. Vaccine 2009, 27, 6420–6423. [Google Scholar] [CrossRef] [PubMed]
- Rubí-Sans, G.; Cano-Torres, I.; Pérez-Amodio, S.; Blanco-Fernandez, B.; Mateos-Timoneda, M.A.; Engel, E. Development and Angiogenic Potential of Cell-Derived Microtissues Using Microcarrier-Template. Biomedicines 2021, 9, 232. [Google Scholar] [CrossRef] [PubMed]
- Mendonça, R.Z.; Prado, J.C.M.; Pereira, C.A. Attachment, spreading and growth of VERO cells on microcarriers for the optimization of large scale cultures. Bioprocess Eng. 1999, 20, 565. [Google Scholar] [CrossRef]
- Luo, X.; Niu, Y.; Fu, X.; Lin, Q.; Liang, H.; Liu, L.; Li, N. Large-Scale Microcarrier Culture of Chinese Perch Brain Cell for Viral Vaccine Production in a Stirred Bioreactor. Vaccines 2021, 9, 1003. [Google Scholar] [CrossRef]
- Ton, C.; Stabile, V.; Carey, E.; Maraikar, A.; Whitmer, T.; Marrone, S.; Afanador, N.L.; Zabrodin, I.; Manomohan, G.; Whiteman, M.; et al. Development and scale-up of rVSV-SARS-CoV-2 vaccine process using single use bioreactor. Biotechnol. Rep. 2023, 37, e00782. [Google Scholar] [CrossRef]
- Wu, S.-C.; Liu, C.-C.; Lian, W.-C. Optimization of microcarrier cell culture process for the inactivated enterovirus type 71 vaccine development. Vaccine 2004, 22, 3858–3864. [Google Scholar] [CrossRef]
- Mendonça, R.Z.; Pereira, C.A. High density VERO cell culture on microcarriers in a cell bioreactor. Bioprocess Eng. 1995, 12, 279. [Google Scholar] [CrossRef]
- Cherry, R.S.; Papoutsakis, E.T. Physical mechanisms of cell damage in microcarrier cell culture bioreactors. Biotechnol. Bioeng. 1988, 32, 1001–1014. [Google Scholar] [CrossRef]
- Maillot, C.; deIsla, N.; Loubiere, C.; Toye, D.; Olmos, E. Impact of microcarrier concentration on mesenchymal stem cell growth and death: Experiments and modeling. Biotechnol. Bioeng. 2022, 119, 3537–3548. [Google Scholar] [CrossRef] [PubMed]
- Nienow, A.W.; Rafiq, Q.A.; Coopman, K.; Hewitt, C.J. A potentially scalable method for the harvesting of hMSCs from microcarriers. Biochem. Eng. J. 2014, 85, 79–88. [Google Scholar] [CrossRef]
- Strobl, F.; Duerkop, M.; Palmberger, D.; Striedner, G. High shear resistance of insect cells: The basis for substantial improvements in cell culture process design. Sci. Rep. 2021, 11, 9413. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.; Lyu, J.; Li, J.; Li, C.; Zhang, Y.; Guo, Y.; Wang, Y.; Zhang, Y.; Chen, K. Application of bioreactor technology for cell culture-based viral vaccine production: Present status and future prospects. Front. Bioeng. Biotechnol. 2022, 10, 921755. [Google Scholar] [CrossRef] [PubMed]
- Loubière, C.; Delafosse, A.; Guedon, E.; Chevalot, I.; Toye, D.; Olmos, E. Dimensional analysis and CFD simulations of microcarrier ‘just-suspended’ state in mesenchymal stromal cells bioreactors. Chem. Eng. Sci. 2019, 203, 464–474. [Google Scholar] [CrossRef]








| 3c | 10c | 50c | 250 L | 1000 L | |
|---|---|---|---|---|---|
| Bioreactor WV * (L) | 1.25–3.75 | 3.3–10 | 18–40 L | 125–250 | 500–1000 |
| Actual WV ** (L) | 2.5 | 10 | 40 | 190 | 900 |
| Volume taken from previous culture (L) | - *** | 2.5 | 10.0 | 40.0 | 190.0 |
| Actual MC Conc. (g/L) | 3.0 | 3.8 | 3.9 | 3.8 | 3.8 |
| Actual cells/bead | 20.0 | 21.7 | 20.7 | 17.9 | 18.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ebrahimian, A.; Schalk, M.; Dürkop, M.; Maurer, M.; Bliem, R.; Kühnel, H. Seed Train Optimization in Microcarrier-Based Cell Culture Post In Situ Cell Detachment through Scale-Down Hybrid Modeling. Bioengineering 2024, 11, 268. https://0-doi-org.brum.beds.ac.uk/10.3390/bioengineering11030268
Ebrahimian A, Schalk M, Dürkop M, Maurer M, Bliem R, Kühnel H. Seed Train Optimization in Microcarrier-Based Cell Culture Post In Situ Cell Detachment through Scale-Down Hybrid Modeling. Bioengineering. 2024; 11(3):268. https://0-doi-org.brum.beds.ac.uk/10.3390/bioengineering11030268
Chicago/Turabian StyleEbrahimian, Atefeh, Mona Schalk, Mark Dürkop, Michael Maurer, Rudolf Bliem, and Harald Kühnel. 2024. "Seed Train Optimization in Microcarrier-Based Cell Culture Post In Situ Cell Detachment through Scale-Down Hybrid Modeling" Bioengineering 11, no. 3: 268. https://0-doi-org.brum.beds.ac.uk/10.3390/bioengineering11030268