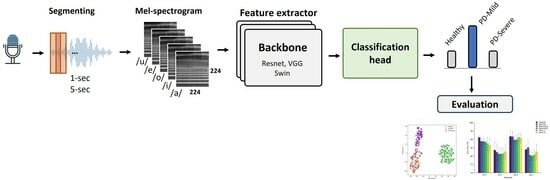
1. Introduction
Parkinson’s disease (PD) is a progressive neurodegenerative disorder characterized by motor symptoms like tremors, rigidity, and slowed movement [
1,
2,
3]. However, pathology underlying PD begin years before the clinical diagnosis, with early manifestations like hyposmia, speech disorders, depression, constipation, and sleep disturbances frequently overlooked [
4,
5]. Diagnosing PD during the initial phase and initiating treatment can potentially impede the rate of progression of this degenerative disorder [
6].
While neurological examination methods like the Movement Disorder Society Unified Parkinson’s Disease Rating Scale (MDS-UPDRS) and brain scans are among the main criteria for diagnosing PD, they have limitations such as cost, accessibility, clinician bias, and difficulty monitoring progression and treatment effectiveness [
1,
2,
3,
7,
8]. Therefore, there is a need for alternative diagnostic approaches that are more objective, cost-effective, and accessible.
Speech difficulties are often one of the initial and most serious signs of PD, severely affecting how patients communicate and their overall quality of life [
9]. Over 80% of PD patients have some vocal dysfunction, including decreased volume, lack of tone, reduced fundamental frequency range, slurred speech, or abnormal rhythms and melodies [
10,
11]. This can occur up to 5 years before motor symptoms like tremors appear [
12,
13]. While assessing writing and walking needs specialized devices, voice can be captured and analyzed without special equipment or clinic visits [
13]. Therefore, speech analysis provides a promising opportunity for early PD detection and continuous monitoring.
Various acoustic analysis techniques including measuring fundamental frequency variation, noise parameters, and non-linear dynamics, have been explored for detecting and quantifying vocal symptoms [
14,
15]. However, recent research has increasingly focused on leveraging advanced machine learning and neural network approaches to automatically detect PD through speech analysis [
16]. Significant work has centered on selecting optimal features for shallow classifiers as well as determining ideal architectures for deep learning classifiers.
The first approach involves hand-crafting acoustic features, including certain variants of the jitter, shimmer, and harmonic-to-noise ratio that are indicative of PD speech impairments [
17,
18,
19,
20,
21,
22] and using traditional machine learning (ML) methods, such as support vector machines (SVM), random forests (RF), k-nearest neighbors (KNN), and regression trees (RT) [
20,
21,
22,
23,
24,
25,
26,
27].
Mamun et al. tested ten algorithms on 195 vocal recordings, finding that LightGBM, a gradient-boosting method, achieved 95% accuracy in classifying PD [
23]. Govindu et al. recently studied early PD detection via telemedicine using ML models on audio data from 30 PD and 30 control subjects. Their RF classifier had the best performance—91.83% accuracy and 0.95 sensitivity for detecting PD [
20]. Wang et al. implemented 12 machine learning models on the 401 voice biomarkers dataset to classify subjects as PD or not. They also built a custom deep learning model with a classification accuracy of 96.45% [
24]. Pramanik et al. achieved high accuracy in PD detection using Naïve Bayes algorithms [
28]. Other studies focused on feature selection techniques. Lamba et al. tested combinations of three selection methods (mutual information gain, extra tree, genetic algorithm) and three classifiers (Naive Bayes, KNN, RF), finding that the genetic algorithm plus RF performed best with 95.58% accuracy [
25].
In contrast to the previous approach, which primarily used manual feature engineering and shallow classifiers, the second approach harnesses deep learning to automatically learn features directly from speech data. Various neural network architectures have been designed and tested, including Convolutional Neural Networks (CNNs), Recurrent Neural Networks (RNN) like Long Short-Term Memory Networks (LSTMs) networks, a combination of them, and more recently, transformer-based models. These models directly learn feature representations from the speech signal or spectrograms, including sustained vowels, continuous speech, and repeating syllables. Deep learning models can alleviate the need for expert-crafted features and have achieved state-of-the-art (SOTA) results on PD detection from speech [
8].
Aversano et al. developed LSTM and CNN models to analyze voice recordings segmented into 1 s intervals consisting of vowels, phrases, and sentences. These voice samples were transformed into mel spectrogram representations as input to the models, which achieved an F1 score of 97%. However, a notable limitation of this study was that the researchers did not ensure that the training and validation sets were speaker-independent, which could potentially introduce biases and may limit the generalizability of the models’ performance [
29]. Similarly, Shah et al. employed a CNN-based model that analyzed 1 s speech chunks transformed into log-scaled mel spectrograms (LMS) for detecting PD from vowel phonations of /a/ and /i/, achieving 90.32% accuracy [
30]. Another study employed a MobileNet CNN model with various types of spectrograms as input. The findings indicated that speech energy spectrograms and mel spectrograms yielded the highest accuracy rates of 96% and 92%, respectively [
31]. A study by Khojasteh et al. evaluated the performance of a CNN model on sustained vowel phonation recordings of the /a/ lasting over 5 s. When tested on 2 s voice samples segmented into 815 ms frames, the CNNs achieved a classification accuracy of 75.7%. An interesting aspect of their approach involved data augmentation techniques like flipping (vertically and horizontally) and rotating the frames, which were applied to the training dataset. However, since the inputs were spectrogram-based images representing time-frequency information, such spatial transformations may not have been suitable augmentation techniques [
8]. Quan et al. employed an end-to-end model incorporating both 2D and 1D CNNs to achieve 92% accuracy in classifying PD based on speech tasks involving the reading of both simple and complex sentences. Their model operated on a sequence of overlapping segments derived from the LMS representation of the input audio. However, the study did not specify the length of this sequence of overlapping segments [
10].
Furthermore, some researchers further improve performance by using transfer learning to adapt these speech models, leveraging knowledge already gained on other tasks. Hireš et al. proposed an ensemble approach involving multiple fine-tuned versions of the Xception deep learning model. When applied to a subset of the sustained vowel recordings dataset (PC-GITA), focusing on the vowels /a/, /i/, /o/, /u/, and /e/, this ensemble method achieved an impressive 99% accuracy in classifying the presence of PD based solely on the voice recordings. In their approach, the 1 s voice signal was transformed into a spectrogram, which was then blurred before being processed by the models [
13]. In another study, Wodzinski et al. fine-tuned a ResNet architecture model using a subset of the PC-GITA dataset containing only the vowel sound /a/. By transforming the audio recordings into spectrograms, their model achieved an accuracy of over 90% in classifying the presence of PD [
11]. More recently, Klempíř et al. found that self-supervised speech models, such as wav2vec which have been pre-trained on 960 h of 16 kHz English speech, generate valuable embeddings for PD detection. These models achieved AUROC (area under the receiver operating characteristic curve) scores ranging from 0.77 to 0.98 across various datasets, which included repeated /pa/ syllables. Notably, this pipeline can be immediately applied to raw audio signal recordings without the need for segmenting [
32]. In summary, the deep learning approach shows promise for PD detection from voice, with recent work achieving accuracies over 90% using techniques like CNNs, LSTM models, and self-supervised learning.
Prior studies have focused on binary classification of PD detection from voice recordings, distinguishing between people with PD and healthy controls. However, clinical applications would benefit from more granular subtype classification beyond this binary distinction [
33]. In this work, we first explored the use of multi-class classification to detect PD and differentiate between various stages based on their MDS-UPDRS III scores. Part III of the MDS-UPDRS assesses motor function in Parkinson’s disease patients. We trained models to classify voice recordings into three classes. This paper also compared three DL architectures widely used in computer vision tasks. The models were trained using LMS representations derived from sustained vowel phonations from a publicly accessible dataset. Secondly, the study examined how the length of audio clips and particular vowel sounds impacted the effectiveness of these models. Additionally, previous studies segmented audio recordings before analysis but did not evaluate model performance on full recordings; in this work, we applied an ensemble method across segments to obtain overall classifications for entire segments after splitting. Finally, we employed visualization techniques such as Grad-CAM [
34] and t-SNE [
35] to provide possible explanations of the deep learning model’s predictions, highlighting discriminative regions in the LMS inputs that influence particular classification decisions.
3. Results and Discussion
In this section, we will describe and discuss our results in detail while evaluating the studied models’ performance.
3.1. Classification Performance
A stratified patient-independent three-fold cross-validation approach was utilized for all experiments, where the data was partitioned into three folds with no patient overlap across folds to avoid data leakage and reduce potential biases in model evaluation. The model was trained on two folds and evaluated on the held-out fold, and this was repeated three times so that each fold served as the evaluation set once. This ensured a rigorous assessment of model performance on unseen data. We decided not to use a separate test set due to the small database size. To mitigate potential issues caused by an imbalance of class distribution, we utilized the train-time oversampling technique to achieve a more balanced class distribution [
51,
52].
The cross-validated performance metrics, including precision, recall, F1 score, and accuracy, for each model are presented in
Table 4 and
Table 5. Additionally,
Figure 7 depicts a graphical representation of the cross-validated classification accuracy for each model. In addition, performance by two additional recent architectures were compared in
Table S2.
Table 4 highlights that utilizing the first 5 s of each recording results in higher classification accuracy across all models compared to using only the initial 1 s segment. While all models demonstrate strong performance in correctly identifying HC subjects, they face challenges distinguishing between varying degrees of PD severity. The FS-5 dataset exhibited superior performance in classifying the different stages of PD. When considering only the recognition of HC subjects, the Swin_s model slightly outperformed other models, demonstrated the best performance in terms of precision (97.00 ± 4.24), recall (100.00 ± 0), and F1 score (98.33 ± 2.36). However, its performance showed minimal deviation compared to the other models.
Our findings indicated that the models demonstrated better accuracy when using longer phonation samples as input. As shown in
Table 5, models trained on complete audio segments, rather than just the initial segment, exhibit higher average accuracy on 5 s datasets (AS-5). However, this improvement comes at the cost of increased performance variability, as evidenced by larger standard deviations. Notably, the Swin Transformer models demonstrate the largest gain of around 3% when utilizing the AS-5 dataset. In contrast, for the 1 s dataset, particularly the ResNet models, there is no improvement when using the AS dataset. Among the tested models, VGG19 experiences the most significant boost on the 1 s dataset when trained on all segments compared to just the initial segments.
Overall, utilizing complete audio clips for training tends to improve model accuracy, especially for longer 5 s datasets, although this benefit is less pronounced on the shorter 1 s dataset (AS-1). In addition, visual inspection of bar plots in
Figure 7 suggests that, for the specific task we have, the deeper architectures do not demonstrate a substantial improvement in accuracy when compared to their shallower counterparts. Furthermore, the transformer-based model showed noticeable performance gains when trained on the AS dataset. Conversely, the CNN-based models evaluated did not exhibit significant improvements from utilizing the full segmented data.
The proposed models for 5 s datasets were evaluated using cumulative confusion matrices and receiver operating characteristic (ROC) curves across three-fold cross-validation. The confusion matrices aggregated results across folds to showcase the overall model performance. Color bars accompanying the confusion matrices illustrated the proportions of observations within each class that were correctly or incorrectly classified, with values ranging from 0 to 1. The ROC curves plotted the trade-off between the true positive rate and the false positive rate, depicting the diagnostic capability of the models. A One versus Rest (OvR) method constructed the ROC curves. The area under the ROC curve (AUC) signified model performance, with higher values indicating better classification ability. Across models, the AUC for the HC class approached 1.00 (
Figure 8 and
Figure 9), demonstrating strong identification of healthy subjects. For PD classes, VGG16 achieved slightly higher AUCs compared to other models. Furthermore, the analysis revealed an increase in the AUC from the FS to the AS dataset, particularly for the PD_Mild class, with a 4% improvement. This suggests that the models exhibited slightly better discrimination capabilities when utilizing the full-segment dataset. Furthermore, the transformer-based models exhibited higher AUC values when trained on the larger AS-5 dataset, suggesting that these models benefited from the increased data availability for improved classification performance.
The analysis of the confusion matrices in
Figure 8 and
Figure 9 suggests that the models excel at accurately identifying samples from the HC group, exhibiting the highest precision and recall for this class. For the FS-5 dataset, there were no instances where an HC sample was incorrectly predicted as PD_Severe or vice versa. However, some instances labeled PD_Severe were misclassified as PD_Mild, and vice versa, indicating potential challenges in distinguishing between these two classes. To better evaluate the VGG16 model’s accuracy for different vowels, we grouped the results by the sustained vowel present in the dataset. The confusion matrices for each vowel are shown in
Figure 10. Of the vowels, /u/ had the highest recall for HC and PD_Severe groups (100%) while having a lower recall value for the PD_Mild group (75%).
Although binary classification was not employed in this study, we combined the results to compare accuracy with previous works that utilized the Italian-speaking Parkinson’s speech dataset. Specifically, we categorized HC as negative and all PD cases as positive. The accuracy results of this binary classification are summarized in
Table 6.
These results are promising; however, recent studies [
53,
54] indicated that the models employed for pathological voice detection are typically trained using small-scale data, hindering their ability to perform consistently across diverse datasets. As a result, the performance of these models fluctuates considerably depending on the dataset encountered. This is largely due to the scarcity and variability in the quality of medical voice recordings available for training such systems [
54]. This can limit model robustness compared to speech recognition systems trained on ample large-scale datasets. For greater generalizability and diagnostic precision, more consistent and substantial medical voice datasets are required.
Table 6.
Comparison of accuracy results obtained on the Parkinson Italian speaking dataset.
Table 6.
Comparison of accuracy results obtained on the Parkinson Italian speaking dataset.
| Author | Model | Accuracy [%] |
|---|
| Aversano et al. [29] | LSTM | 97.1 |
| Klempíř et al. [32] | Wav2Vec | 95.0 |
| Hireš et al. [54] | Xception | 97.8 |
| Toye et al. [17] | SVM | 98.9 1 |
| Current study | Swin_s | 98.5 ± 2.50 |
| Current study | VGG16 | 98.1 ± 3.23 |
In previous studies [
11,
29] on PD classification using audio recordings, researchers have typically segmented the recordings into smaller parts before extracting features and training machine learning models. The researchers assessed the models’ performance on the segmented audio excerpts and reported the corresponding results for these segments. However, they did not provide performance results for complete audio samples. This study employed a simple ensemble method to enable a fair evaluation and comparison of different audio segmentation approaches. Specifically, we passed each segment through the trained model to get a prediction, then took the most common predicted class across all segments as the final prediction for the recording, effectively using majority voting. This allows the comparison of different segmenting approaches equally in terms of overall recording classification. After using this approach, we calculated the cumulative confusion matrix and accuracy, as shown in
Figure 11 for the AS-5 dataset. This is a more realistic test scenario, as in real-world applications, we would need to make predictions on individual audio. When implementing this approach, the accuracy of the VGG19 model increased by around 1% compared to results on the AS-5 dataset. Accuracy for the other models did not change significantly or even decreased slightly for this dataset. Despite overall lower performance compared to not using ensembling, our dataset still achieved slightly higher accuracy than when we used the FS dataset, especially when leveraging transformer-based models. This increases more pronounce for the AS-1 dataset that is shown in
Figure S1.
We further evaluated the segmentation and ensemble approach by applying it separately to each individual vowel sound, aiming to determine which vowel benefited the most from this technique and achieved the highest performance. The results summarized in
Table 7 and
Table 8 demonstrate that the vowels /u/ and /o/ may have the greatest ability among the models to differentiate between Parkinson’s classes. Notably, the findings suggest that when utilizing solely the vowel /u/ for classification with the VGG16 model, an impressive F1 score of 96% can be attained. The performance on vowel /u/ in [
29] contributed to the overall improved accuracy across the different methods utilized. These results align with earlier findings in [
55] that the vowel /u/ had the highest classification accuracy out of the vowels /a/, /o/ and /u/ tested. Rusz et al. [
15] provided further support, identifying abnormalities in vowel articulation and acoustics, such as reduced vowel space area, among PD patients, especially for the vowel /u/.
3.2. Grad Cam Feature Visualization
Grad-CAM (Gradient-weighted Class Activation Mapping) is a visual explanation technique for CNNs [
34]. Grad-CAM utilizes the gradient information from the final convolutional layer of a CNN to generate a heat map representing the regions of the input image that are most relevant for the network’s prediction. Specifically, it computes the gradients of the target concept (i.e., the class output) with respect to the feature maps of the last convolutional layer. By pooling these gradients over the spatial dimensions, Grad-CAM produces a coarse localization map that highlights the parts of the image that have the greatest influence on CNN’s decision [
34,
38]. The architecture explaining the Grad-CAM technique is shown in
Figure S2.
The Grad-CAM feature map visualizations in
Figure 12 represent three 5 s audio clips of the vowel sound /o/ from the FS-5 dataset. To maintain consistency, we exclusively used data from the second fold of the FS-5 dataset and the corresponding trained models.
The generated heatmaps highlighted the specific regions in an LMS input image that significantly influenced the model’s prediction. A comparison of the visualization results across different columns revealed key differences between the CNN-based and Swin transformer-based architectures. The CNN models demonstrated more localized attention, focusing on specific local areas in the images [
56]. In contrast, the visualizations for the Swin transformer network displayed attention that was more scattered and less spatially localized.
The models generally placed less emphasis on the higher frequency components of the LMSs, particularly in the range greater than 1024 Hz, suggesting that these regions were less discriminative for the classification task. However, it was noteworthy that the Swin Transformer models, in addition to their focus on lower frequencies, less than 512 Hz, also exhibited sensitivity to relatively higher frequencies when detecting healthy control subjects. Furthermore, the ResNet 18 model for the healthy control class demonstrated primary activation in the high-frequency range.
When examining the temporal patterns for the healthy class, it was evident that CNN models primarily focused on the first half to the middle of the audio clips, while transformer-based models were more consistent across time frames. For the mild class, models generally concentrated on the middle period. For the severe class, VGG16 displayed a distinct pattern compared to the other studied models. This model was activated on the middle frequency range (around 2048 Hz) and the timeframes of the initial segments. Additionally, there was a moderately intense region towards the end of the spectrogram. In contrast, the other models focused more on the second half of the audio clips and lower frequencies.
This suggests that the network heavily relies on the spectral patterns in this specific time-frequency region, indicating that the network is also considering some higher-frequency components.
3.3. Analyzing Feature Extraction Capability
In the previous section, Grad-CAM visualizations demonstrated qualitative differences between the features extracted by different architectures on our classification FS-5 dataset. To further analyze these representations, the t-distributed Stochastic Neighbor Embedding (t-SNE) technique can be utilized to project high-dimensional feature spaces into a 2D representation, allowing for visualization and interpretation of the learned representations.
Figure 13 presents 2D scatter plots that visualize the distribution of features extracted from the layer just before the classifier in each model. Each class is represented by a different color, allowing for visual analysis of how well the features separate the classes prior to classification.
The t-SNE visualization clearly shows three distinct clusters corresponding to the Healthy, PD_Mild, and PD_Severe classes across all models. Architectures like VGG16, Swin_s, and ResNet50 exhibit cleaner separations between these class clusters, suggesting their ability to extract more discriminative features from the log mel spectrogram images. Notably, the ResNet50 model forms the most compact clusters, indicating higher feature similarity within each class. However, there is some overlap between the PD_Mild and PD_Severe classes, particularly in the region where their feature points intersect. This overlap suggests that certain mild and severe cases may share similar feature characteristics, making it challenging to distinguish them based solely on the extracted features.
Despite the subtle overlap between PD_Mild and PD_Severe classes, all models successfully separated the Healthy class from the Parkinson’s disease classes, demonstrating the effectiveness of using log mel spectrogram images for distinguishing between healthy and Parkinson’s voices.
4. Conclusions
This study explored multi-class classification of Parkinson’s disease from speech recordings using deep learning approaches. Several popular CNN and transformer models were trained on log mel spectrogram representations of sustained vowel recordings to categorize samples as healthy controls, mild, or severe Parkinson’s disease labeled based on their MDS-UPDRS III scores. The models demonstrated strong capabilities to distinguish healthy samples from those with Parkinson’s, achieving over 95% precision. However, they struggled to reliably differentiate between mild and severe Parkinson’s, with classification precision closer to 85%. The findings revealed that models performed better when utilizing longer speech segments. The Swin transformer architecture attained the best accuracy in terms of binary classification, though its superiority over CNNs was marginal for this task. Considering overall accuracy, VGG16 can be proposed as the best model with 91.8%. Applying ensemble techniques across segments and focusing analysis on vowels, /u/ and /o/ recordings further improved accuracy by 1–4%. Moreover, visualization methods highlighted discriminative regions and features learned by models, showing transformers identify more widespread patterns while CNNs focus on localized spectrogram areas.
A key limitation of this study was the relatively small dataset size, which may have impacted the models’ ability to reliably distinguish between mild and severe cases of Parkinson’s disease. The limited availability of large-scale, well-annotated medical datasets can hinder the generalization capabilities of such models for real-world clinical applications.
In conclusion, this work demonstrates the potential of leveraging deep learning techniques on spectrogram inputs derived from voice recordings to enable non-invasive detection and monitoring of different stages of Parkinson’s disease progression. However, to further enhance the identification of disease severity from patient voices, our future work will focus on building larger multi-class labeled datasets of Parkinson’s cases. Additionally, further research could explore a broader range of SOTA architectures and input representations beyond log mel spectrograms, potentially enhancing the classification accuracy.