1.1. Biological Background
Caenorhabditis elegans is one of the most important invertebrate model organisms in biological research. It has the characteristics of a short life cycle, a simple physiological structure, and a transparent worm body which makes for easy observation. Since the early 1960s, it has been widely used as a popular model organism [
1]. Its research spans multiple disciplines, including large-scale gene function and characterization research [
2], the complete lineage tracing of whole-body cells, the structural construction of the animal nervous system connection group [
3], etc. Adult
C. elegans are about 1 mm long and live in the soil. Under normal conditions, most are hermaphrodites and a few are males. The proportion of males can be greatly increased under special circumstances.
C. elegans can grow and reproduce at 12 to 25 °C. In an environment of 25 °C, most
C. elegans have an average lifespan of 12.1 days, with a standard deviation of 2.3 days of incubation [
4].
C. elegans also provides an ideal model for studying the variability inducement that leads to differences in individual health and lifespan: the relative variability reflected in the lifespan cycle of about two weeks is almost as great as that of human beings from birth to 80 years old. In recent years, with the application of cutting-edge technologies, such as machine learning and artificial intelligence, in biological research, many researchers have used methods such as deep learning in biological research. Hua et al. developed an end-to-end ECG classification algorithm to help classify ECG signals and reduce the workload of physicians [
5]. He et al. developed a novel evolvable adversarial framework for COVID-19 infection segmentation [
6]. Mu et al. proposed a progressive global perception and local polishing (PCPLP) network to automatically segment pneumonia infection caused by COVID-19 in computed tomography (CT) images [
7]. Zhao et al. developed a deep learning model combining a feature pyramid with a U-Net++ model for the automatic segmentation of coronary arteries in ICA [
8]. Liu et al. proposed a new method for nuclei segmentation [
9]. The proposed method performs end-to-end segmentation of pathological tissue sections using a deep fully convolutional neural network. Cao and Liu proposed a method for segmenting the terminal bulb of
C. elegans based on the U-Net network [
10]. This method solves the problem encountered with traditional single-stage networks that are not suitable for small samples. Lin et al. presented a quantitative method for measuring physiological age
in C. elegans using a convolutional neural network (CNN) [
11]. Fudickar et al. proposed an image acquisition system [
12]. This system was used to create large datasets containing entire dishes of
C. elegans. At the same time, the authors used the object detection framework Mask R-CNN to localize, classify, and predict the outlines of nematodes.
Regarding the lifespan assessment of
C. elegans, there are currently two main research directions: one is to use physiological changes for evaluation; the other is to use biomarkers for evaluation. The physiological changes here refer to the physiological changes in
C. elegans that can be directly observed, such as the swallowing rate of the pharynx, measurement of image entropy, measurement of appearance, measurement of exercise capacity, and measurement of auto-fluorescence. In the study of Zhang et al., based on the view that the aging of
C. elegans is a plasticity process discovered by predecessors, a large number of comparative experiments were carried out on the aging of
C. elegans [
13]. Experiments have found that there is a big difference in lifespan among individuals of
C. elegans. Short-lived
C. elegans have a lifespan of about 12 days, while long-lived
C. elegans can survive for more than 20 days. Through more detailed experiments, the authors determined the difference between the long-lived and short-lived
C. elegans in the physiological process of aging. The difference in the lifespan of
C. elegans is mainly concentrated in the time period between reproductive maturity and death, which is less related to the time of larval growth. Although it takes an average of 2.1 days for larvae to develop (accounting for 17.3% of the average lifespan), the variability in the development time is less than 0.1% of their total lifespan. In the subsequent lifespan process, the physiological health status and the rates of physiological health changes are related to the total lifespan of
C. elegans. At the same time, it is also to be pointed out that the differences in physiological changes between different
C. elegans at the same stage are much smaller than the differences in physiological changes between different
C. elegans at the same absolute time. Stroustrup et al. designed a set of lifespan machines to judge and predict the lifespan of
C. elegans by detecting the movement status and movement ability of
C. elegans and achieved better results [
14]. Martineau et al. extracted hundreds of morphological, postural, and behavioral features from
C. elegans activity videos and used support vector machines (SVM) to analyze their direct relationship with
C. elegans lifespan [
15]. Lin et al. presented quantitative methods to measure the physiological age of
C. elegans with convolution neural networks (CNNs), which measured ages with a granularity of days and achieved a mean absolute error (MAE) of less than 1 day [
11]. Furthermore, they proposed two models: one was based on linear regression analysis and the other was based on logistic regression. The linear-regression-based model achieved a test MAE of 0.94 days, while the logistic-regression-based model achieved an accuracy of 84.78 percent with an error tolerance of 1 day. The advantage of using physiological changes for assessment is that this method has a higher accuracy rate and is applicable to various
C. elegans mutants. However, because the research is limited to the
C. elegans body and lacks the possibility of technological migration, the significance for human research is relatively limited.
Compared with physiological changes, biomarkers mainly consist of life-related genes or microRNA promoters carrying fluorescent proteins. The relevant signal pathway mechanism behind the genes is clear, and there is the possibility of technology migration, which has potential guiding significance for the assessment of human aging [
16]. For example, Wan et al. designed a
C. elegans life prediction algorithm based on Naive Bayes [
17]. They investigated the relationship between
C. elegans gene sequencing information, protein expression information, and lifespan. They proposed a feature selection method based on Naive Bayes to predict the effects of
C. elegans genes on biological life. However, obtaining gene sequencing information and protein expression information for
C. elegans leads to the death of the worms, which is not conducive to the verification of the test set and the further development of research. At the same time, the cost of extracting gene sequencing information from
C. elegans is expensive, and it is not suitable for repeated experiments. Saberi-bosari et al. also selected biomarkers as the research objects, in order to use the Mask R-CNN [
18] algorithm to identify the neurodegenerative sub-cellular processes that appear after the senescence of the
C. elegans PVD neurons, and they used this information to determine current lifespan stages of
C. elegans [
19]. The biological state was divided into three states: young and old adults, cold-shocked and non-shocked nematodes, and cold-shocked and aged worms. Finally, a classification accuracy of 85% was obtained. However, in actual research, it has been found that the currently used biomarkers have the following two problems. On the one hand, the overall performance of biomarkers is relatively poor, which may be due to the limited influence of a single gene on lifespan [
20]. On the other hand, some endogenous genes have a certain evaluative power in the wild type, but they often have poor evaluative potential in specific mutant strains (such as daf-16). This is because the genes used in the evaluation are often limited to specific signaling pathways, and the phenomenon of aging is jointly regulated by multiple signaling pathways [
21].
We selected proteostasis as a life-related indicator because most biological activities are dependent on protein function and many life-related signal pathways in
C. elegans show the regulation of proteostasis. With the aging of
C. elegans, protein accumulation will gradually increase [
22]. At the same time, in the process of human aging, proteostasis is also related to many senile diseases [
23], such as Alzheimer’s disease [
24], Parkinson’s disease [
25], and so on. Intrinsic protein aggregation is a biomarker of aging. Knowing how to regulate it will help understand the underlying mechanisms of aging and protein aggregation diseases [
26]. For the detection of proteostasis imbalance, there are also mature visual biomarkers in
C. elegans metastable protein [
27]. In summary, the physiological index of proteostasis change is closely related to aging, involves multiple signaling pathways, is conservative among species, and is easy to detect, so it is a good candidate for lifespan estimation. To minimize the impact on
C. elegans, the response is closest to the aging process in the natural state. We selected firefly luciferase protein, which has not been reported as being related to pathological processes associated with a variety of metastable proteins.
C. elegans carrying multiple copies of the firefly luciferase gene will not exhibit premature aging and paralysis phenotypes. This is the first study to use protein aggregation as a biomarker to estimate the lifespan of
C. elegans.
1.2. Convolutional Network Background
In recent years, neural networks, representing an emerging deep learning method, have continuously enabled breakthroughs in various fields. Among them, convolutional neural networks (CNNs) [
28], as deep feed-forward neural networks involving convolutional calculations, can better obtain spatial position and shape information from an image. They are widely used in many fields, including object tracking, pose estimation, text detection and recognition, visual saliency detection, action recognition, and scene labeling.
Generally, a convolutional neural network consists of a convolutional layer, a pooling layer, and a fully connected layer. The input image is respectively processed by the convolutional layer, the pooling layer, and the fully connected layer, and the feature map is finally outputted. Limited by computer performance and the acquisition of datasets, the convolutional neural network model was unutilized for decades, until the proposal of AlexNet [
29] in 2012, which stimulated the frenzy of convolutional neural network research. In this study, we chose VGG16 [
30], Inceptionv3 [
31], MobileNetV3 [
32], ResNet50 [
13], and DenseNet [
33] as candidate benchmark CNN models.
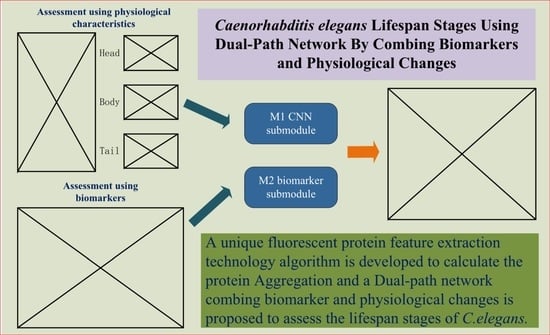
Given the shortcomings of the above two methods, we hope to design an evaluation and prediction method to maximize accuracy and have better mobility for future research on the human lifespan. To this end, in this paper, we propose an estimation method for C. elegans lifespan stages combining a C. elegans biomarker (protein aggregation) and a high-efficiency attention improved network (Att-efficientNet). Compared to using shape and motion trajectory features, this is a new attempt. The dual-path feature fusion model based on deep neural networks can extract local features of C. elegans through neural networks and compensate for the loss of global features by calculating fluorescent protein aggregation information and finally output the multi-stage classification results for C. elegans lifespan stages. In this paper, we divide the lifespan of C. elegans into six stages. This method has basically met the needs of biological research to predict the lifespan stages of C. elegans. This work makes several main contributions:
The study is the first to estimate the lifespan stages of C. elegans using the six-stage standard from fluorescence microscope images;
A dual-path network combining biomarker and physiological changes is proposed to jointly estimate the lifespan stages of C. elegans from fluorescence microscope images;
An Att-EfficientNet was developed to extract physiological changes and a unique fluorescent protein feature extraction technology was developed to calculate protein aggregation degrees;
We evaluated the proposed method on a dataset with 4593 fluorescence microscope images of C. elegans and achieved promising results in lifespan stage estimation compared with several other machine learning methods.