Long Non-Coding RNAs as Emerging Regulators of Pathogen Response in Plants
Abstract
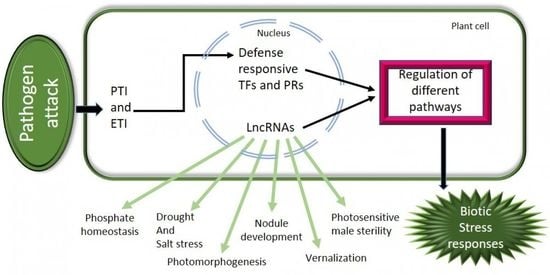
:1. Introduction
2. Origin and Database Development of Plant lncRNAs
3. Involvement of lncRNAs in Various Biological Processes
4. Roles of lncRNAs in Various Biotic Stress Responses
4.1. Long Non-Coding RNAs against Fungal Infection
4.2. Long Non-Coding RNAs against Viral Infection
4.3. Long Non-Coding RNAs against Bacterial Infection
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Budak, H.; Kaya, S.B.; Cagirici, H.B. Long Non-coding RNA in Plants in the Era of Reference Sequences. Front. Plant Sci. 2020, 11, 276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Urquiaga, M.C.O.; Thiebaut, F.; Hemerly, A.S.; Ferreira, P.C.G. From Trash to Luxury: The Potential Role of Plant LncRNA in DNA Methylation during Abiotic Stress. Front. Plant Sci. 2021, 11, 603246. [Google Scholar] [CrossRef]
- Tyagi, S.; Sharma, A.; Upadhyay, S.K. Role of next-generation RNA-seq data in discovery and characterization of long non-coding RNA in plants. In Next Generation Plant Breeding; Intech Open: London, UK, 2018. [Google Scholar] [CrossRef] [Green Version]
- Ponting, C.P.; Oliver, P.L.; Reik, W. Evolution and functions of long noncoding RNAs. Cell 2009, 136, 629–641. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Fan, X.; Lin, F.; He, G.; Terzaghi, W.; Zhu, D.; Deng, X.W. Arabidopsis noncoding RNA mediates control of photomorphogenesis by red light. Proc. Natl. Acad. Sci. USA 2014, 111, 10359–10364. [Google Scholar] [CrossRef] [Green Version]
- Fan, Y.; Yang, J.; Mathioni, S.M.; Yu, J.; Shen, J.; Yang, X.; Wang, L.; Zhang, Q.; Cai, Z.; Xu, C.; et al. PMS1T, producing Phased small-interfering RNAs, regulates photoperiod sensitive male sterility in rice. Proc. Natl. Acad. Sci. USA 2016, 113, 15144–15149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Hu, W.; Hao, J.; Lv, S.; Wang, C.; Tong, W.; Wang, Y.; Wang, Y.; Liu, X.; Ji, W. Genome-wide identification and functional prediction of novel and fungi-responsive lincRNAs in Triticum aestivum. BMC Genom. 2016, 17, 238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, X.; Zheng, H.; Li, J.; Liu, L.; Zhang, X.; Sui, N. Comparative transcriptome analysis reveals new lncRNAs responding to salt stress in sweet sorghum. Front. Bioeng. Biotechnol. 2020, 8, 331. [Google Scholar] [CrossRef] [Green Version]
- Upadhyay, S.K. Long Noncoding RNAs in Plants: Roles in Development and Stress; Academic Press: Cambridge, MA, USA, 2021. [Google Scholar] [CrossRef]
- Pachnis, V.; Belayew, A.; Tilghman, S.M. Locus unlinked to alpha-fetoprotein under the control of the murine raf and Rif genes. Proc. Natl. Acad. Sci. USA 1984, 81, 5523–5527. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Guo, H.; Hu, W.; Ji, W. The Emerging Role of Long Non-Coding RNAs in Plant Defense against Fungal Stress. Int. J. Mol. Sci. 2020, 21, 2659. [Google Scholar] [CrossRef]
- Dinger, M.E.; Pang, K.C.; Mercer, T.R.; Crowe, M.L.; Grimmond, S.M.; Mattick, J.S. NRED: A database of long noncoding RNA expression. Nucleic Acids Res. 2009, 37, D122–D126. [Google Scholar] [CrossRef] [Green Version]
- Wierzbicki, A.T.; Haag, J.R.; Pikaard, C.S. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 2008, 135, 635–648. [Google Scholar] [CrossRef] [Green Version]
- Kung, J.T.; Colognori, D.; Lee, J.T. Long noncoding RNAs: Past, present, and future. Genetics 2013, 193, 651–669. [Google Scholar] [CrossRef] [Green Version]
- Mattick, J.S.; Rinn, J.L. Discovery and annotation of long noncoding RNAs. Nat. Struct. Mol. Biol. 2015, 22, 5–7. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Yang, L.; Chen, L.L. The Diversity of Long Noncoding RNAs and Their Generation. Trends Genet. 2017, 33, 540–552. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.C.; Chang, H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, J.; Liu, J.; Wang, H.; Wong, L.; Chua, N.-H. PLncDB: Plant long non-coding RNA database. Bioinformatics 2013, 29, 1068–1071. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Wang, X.; Deng, W.; Fan, X.; Liu, T.-T.; He, G.; Chen, R.; Terzaghi, W.; Zhu, D.; Deng, X.W. Genomic features and regulatory roles of intermediate-sized non-coding RNAs in Arabidopsis. Mol. Plant 2014, 7, 514–527. [Google Scholar] [CrossRef] [Green Version]
- Heo, J.B.; Sung, S. Vernalization-Mediated Epigenetic Silencing by a Long Intronic Noncoding RNA. Science 2011, 331, 76–79. [Google Scholar] [CrossRef] [Green Version]
- Shin, J.-H.; Chekanova, J.A. Arabidopsis RRP6L1 and RRP6L2 function in FLOWERING LOCUS C silencing via regulation of antisense RNA synthesis. PLoS Genet. 2014, 10, e1004612. [Google Scholar] [CrossRef]
- Van Dijk, E.L.; Chen, C.L.; d’Aubenton-Carafa, Y.; Gourvennec, S.; Kwapisz, M.; Roche, V.; Bertrand, C.; Silvain, M.; Legoix-Né, P.; Loeillet, S.; et al. XUTs are a class of Xrn1-sensitive antisense regulatory non-coding RNA in yeast. Nature 2011, 475, 114–117. [Google Scholar] [CrossRef] [PubMed]
- Schulz, D.; Schwalb, B.; Kiesel, A.; Baejen, C.; Torkler, P.; Gagneur, J.; Soeding, J.; Cramer, P. Transcriptome surveillance by selective termination of noncoding RNA synthesis. Cell 2013, 155, 1075–1087. [Google Scholar] [CrossRef] [Green Version]
- Fox, M.J.; Gao, H.; Smith-Kinnaman, W.R.; Liu, Y.; Mosley, A.L. The exosome component Rrp6 is required for RNA polymerase II termination at specific targets of the Nrd1-Nab3 pathway. PLoS Genet. 2015, 11, e1004999. [Google Scholar] [CrossRef] [Green Version]
- Flynn, R.A.; Almada, A.E.; Zamudio, J.R.; Sharp, P.A. Antisense RNA polymerase II divergent transcripts are P-TEFb dependent and substrates for the RNA exosome. Proc. Natl. Acad. Sci. USA 2011, 108, 10460–10465. [Google Scholar] [CrossRef] [Green Version]
- Hetzel, J.; Duttke, S.H.; Benner, C.; Chory, J. Nascent RNA sequencing reveals distinct features in plant transcription. Proc. Natl. Acad. Sci. USA 2016, 113, 12316–12321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Necsulea, A.; Soumillon, M.; Warnefors, M.; Liechti, A.; Daish, T.; Zeller, U.; Baker, J.C.; Grützner, F.; Kaessmann, H. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature 2014, 505, 635–640. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Wang, J.; Li, Y.; Song, T.; Wu, Y.; Fang, S.; Bu, D.; Li, H.; Sun, L.; Pei, D.; et al. NONCODEV6: An updated database dedicated to long non-coding RNA annotation in both animals and plants. Nucleic Acids Res. 2021, 49, D165–D171. [Google Scholar] [CrossRef]
- Bhatia, G.; Goyal, N.; Sharma, S.; Upadhyay, S.K.; Singh, K. Present Scenario of Long Non-Coding RNAs in Plants. Non-Coding RNA 2017, 3, 16. [Google Scholar] [CrossRef] [Green Version]
- Yi, X.; Zhang, Z.; Ling, Y.; Xu, W.; Su, Z. PNRD: A plant non-coding RNA database. Nucleic Acids Res. 2015, 43, D982–D989. [Google Scholar] [CrossRef] [Green Version]
- Paytuví Gallart, A.; Hermoso Pulido, A.; Anzar Martínez de Lagrán, I.; Sanseverino, W.; Aiese Cigliano, R. GREENC: A Wiki-based database of plant lncRNAs. Nucleic Acids Res. 2016, 4, D1161–D1166. [Google Scholar] [CrossRef]
- Lamesch, P.; Berardini, T.Z.; Li, D.; Swarbreck, D.; Wilks, C.; Sasidharan, R.; Muller, R.; Dreher, K.; Alexander, D.L.; Garcia-Hernandez, M.; et al. The Arabidopsis Information Resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res. 2012, 40, D1202–D1210. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.-Y.; Krishnakumar, V.; Chan, A.P.; Thibaud-Nissen, F.; Schobel, S.; Town, C.D. Araport11: A complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 2017, 89, 789–804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, D.; Yuan, C.; Zhang, J.; Zhang, Z.; Bai, L.; Meng, Y.; Chen, L.-L.; Chen, M. PlantNATsDB: A comprehensive database of plant natural antisense transcripts. Nucleic Acids Res. 2012, 40, D1187–D1193. [Google Scholar] [CrossRef] [PubMed]
- Szcześniak, M.W.; Rosikiewicz, W.; Makałowska, I. CANTATAdb: A collection of plant long non-coding RNAs. Plant Cell Physiol. 2016, 57, e8. [Google Scholar] [CrossRef] [Green Version]
- Kumar, K.; Chakraborty, S. Roles of long non-coding RNAs in plant virus interactions. J. Plant Biochem. Biotechnol. 2021, 30, 684–697. [Google Scholar] [CrossRef]
- Liu, J.; Jung, C.; Xu, J.; Wang, H.; Deng, S.; Bernad, L.; Arenas-Huertero, C.; Chua, N.H. Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell 2012, 24, 4333–4345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, S.; Taneja, M.; Tyagi, S.; Singh, K.; Upadhyay, S.K. Survey of high throughput RNA-seq data reveals potential roles for lncRNAs during development and stress response in bread wheat. Front. Plant Sci. 2017, 8, 1019. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.C.; Liao, J.Y.; Li, Z.Y.; Yu, Y.; Zhang, J.P.; Li, Q.F.; Qu, L.H.; Shu, W.S.; Chen, Y.Q. Genome-wide screening and functional analysis identify a large number of long noncoding RNAs involved in the sexual reproduction of rice. Genome Biol. 2014, 15, 512. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Eichten, S.R.; Shimizu, R.; Petsch, K.; Yeh, C.T.; Wu, W.; Chettoor, A.M.; Givan, S.A.; Cole, R.A.; Fowler, J.E.; et al. Genome-wide discovery and characterization of maize long non-coding RNAs. Genome Biol. 2014, 15, R40. [Google Scholar] [CrossRef] [Green Version]
- Wen, J.; Parker, B.J.; Weiller, G.F. In silico identification and characterization of mRNA-like noncoding transcripts in Medicagotruncatula. Silico Biol. 2007, 7, 485–505. [Google Scholar]
- Bhatia, G.; Sharma, S.; Upadhyay, S.K.; Singh, K. Long Non-coding RNAs Coordinate Developmental Transitions and Other Key Biological Processes in Grapevine. Sci. Rep. 2019, 9, 3552. [Google Scholar] [CrossRef] [Green Version]
- Datta, R.; Paul, S. Long non-coding RNAs: Fine-tuning the developmental responses in plants. J. Biosci. 2019, 44, 77. [Google Scholar] [CrossRef]
- Shafiq, S.; Li, J.; Sun, Q. Functions of plants long non-coding RNAs. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 2016, 1859, 155–162. [Google Scholar] [CrossRef]
- Liu, F.; Marquardt, S.; Lister, C.; Swiezewski, S.; Dean, C. Targeted 3’ processing of antisense transcripts triggers Arabidopsis FLC chromatin silencing. Science 2010, 327, 94–97. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Csorba, T.; Skourti-Stathaki, K.; Proudfoot, N.J.; Dean, C. R-loop stabilization represses antisense transcription at the Arabidopsis FLC locus. Science 2013, 340, 619–621. [Google Scholar] [CrossRef] [Green Version]
- Gultyaev, A.P.; Roussis, A. Identification of conserved secondary structures and expansion segments in enod40 RNAs reveals new enod40 homologues in plants. Nucleic Acids Res. 2007, 35, 3144–3152. [Google Scholar] [CrossRef]
- Ariel, F.; Jegu, T.; Latrasse, D.; Romero-Barrios, N.; Christ, A.; Benhamed, M.; Crespi, M. Noncoding transcription by alternative RNA polymerases dynamically regulates an auxin-driven chromatin loop. Mol. Cell 2014, 55, 383–396. [Google Scholar] [CrossRef] [Green Version]
- Kakar, K.; Zhang, H.; Scheres, B.; Dhonukshe, P. Retraction: CLASP-mediated cortical microtubule organization guides PIN polarization axis. Nature 2014, 508, 274. [Google Scholar] [CrossRef] [Green Version]
- Franco-Zorrilla, J.M.; Valli, A.; Todesco, M.; Mateos, I.; Puga, M.I.; Rubio-Somoza, I.; Leyva, A.; Weigel, D.; García, J.A.; Paz-Ares, J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 2007, 39, 1033–1037. [Google Scholar] [CrossRef]
- Jiao, Y.; Lau, O.S.; Deng, X.W. Light-regulated transcriptional networks in higher plants. Nat. Rev. Genet. 2007, 8, 217–230. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Lu, Q.; Ouyang, Y.; Mao, H.; Zhang, P.; Yao, J.; Xu, C.; Li, X.; Xiao, J.; Zhang, Q. A long noncoding RNA regulates photoperiod-sensitive male sterility, an essential component of hybrid rice. Proc. Natl. Acad. Sci. USA 2012, 109, 2654–2659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, H.; Liu, Q.; Li, J.; Jiang, D.; Zhou, L.; Wu, P.; Lu, S.; Li, F.; Zhu, L.; Liu, Z.; et al. Photoperiod- and thermo-sensitive genic male sterility in rice are caused by a point mutation in a novel noncoding RNA that produces a small RNA. Cell Res. 2012, 22, 649–660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nejat, N.; Mantri, N. Emerging roles of long non-coding RNAs in plant response to biotic and abiotic stresses. Crit. Rev. Biotechnol. 2018, 38, 93–105. [Google Scholar] [CrossRef]
- Seo, J.S.; Sun, H.-X.; Park, B.S.; Huang, C.-H.; Yeh, S.-D.; Jung, C.; Chua, N.-H. ELF18-INDUCED LONG-NONCODING RNA Associates with Mediator to Enhance Expression of Innate Immune Response Genes in Arabidopsis. Plant Cell 2017, 29, 1024–1038. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Liu, Y.; Li, L.; Li, D.; Zhang, Q.; Guo, Y.; Wang, S.; Zhong, C.; Huang, H. Whole transcriptome sequencing of Pseudomonas syringae pv. actinidiae-infected kiwifruit plants reveals species-specific interaction between long non-coding RNA and coding genes. Sci. Rep. 2017, 7, 4910. [Google Scholar] [CrossRef]
- Yu, Y.; Zhou, Y.F.; Feng, Y.Z.; He, H.; Lian, J.P.; Yang, Y.W.; Lei, M.Q.; Zhang, Y.C.; Chen, Y.Q. Transcriptional landscape of pathogen-responsive lncRNAs in rice unveils the role of ALEX1 in jasmonate pathway and disease resistance. Plant Biotechnol. J. 2020, 8, 679–690. [Google Scholar] [CrossRef] [Green Version]
- Xin, M.; Wang, Y.; Yao, Y.; Song, N.; Hu, Z.; Qin, D.; Xie, C.; Peng, H.; Ni, Z.; Sun, Q. Identification and characterization of wheat long non-protein coding RNAs responsive to powdery mildew infection and heat stress by using microarray analysis and SBS sequencing. BMC Plant Biol. 2011, 11, 61. [Google Scholar] [CrossRef] [Green Version]
- Joshi, R.K.; Megha, S.; Basu, U.; Rahman, M.H.; Kav, N.N. Genome Wide Identification and Functional Prediction of Long Non-Coding RNAs Responsive to Sclerotinia sclerotiorum Infection in Brassica napus. PLoS ONE 2016, 11, e0158784. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Q.H.; Stephen, S.; Taylor, J.; Helliwell, C.A.; Wang, M.B. Long noncoding RNAs responsive to Fusarium oxysporum infection in Arabidopsis thaliana. New Phytol. 2014, 201, 574–584. [Google Scholar] [CrossRef]
- Zhang, H.; Chen, X.; Wang, C.; Xu, Z.; Wang, Y.; Liu, X.; Kang, Z.; Ji, W. Long non-coding genes implicated in response to stripe rust pathogen stress in wheat (Triticum aestivum L.). Mol. Biol. Rep. 2013, 40, 6245–6253. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yu, W.; Yang, Y.; Li, X.; Chen, T.; Liu, T.; Ma, N.; Yang, X.; Liu, R.; Zhang, B. Genome-wide analysis of tomato long non-coding RNAs and identification as endogenous target mimic for microRNA in response to TYLCV infection. Sci. Rep. 2015, 5, 16946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Y.; Zhang, H.; Fan, M.; He, Y.; Guo, P. Genome-wide identification of long non-coding RNAs and circular RNAs reveal their ceRNA networks in response to cucumber green mottle mosaic virus infection in watermelon. Arch. Virol. 2020, 165, 1177–1190. [Google Scholar] [CrossRef]
- Bhogireddy, S.; Mangrauthia, S.K.; Kumar, R.; Pandey, A.K.; Singh, S.; Jain, A.; Budak, H.; Varshney, R.K.; Kudapa, H. Regulatory non-coding RNAs: A new frontier in regulation of plant biology. Funct. Integr. Genom. 2021, 21, 313–330. [Google Scholar] [CrossRef]
- Cui, J.; Luan, Y.; Jiang, N.; Ba, H.; Meng, J. Comparative transcriptome analysis between resistant and susceptible tomato allows the identification of lncRNA16397 conferring resistance to Phytophthora infestans by co-expressing glutaredoxin. Plant J. 2017, 89, 577–589. [Google Scholar] [CrossRef] [Green Version]
- Rouhier, N.; Lemaire, S.D.; Jacquot, J.P. The role of glutathione in photosynthetic organisms: Emerging functions for glutaredoxins and glutathionylation. Annu. Rev. Plant Biol. 2008, 59, 143–166. [Google Scholar] [CrossRef]
- Bhatia, G.; Upadhyay, S.K.; Upadhyay, A.; Singh, K. Investigation of long non-coding RNAs as regulatory players of grapevine response to powdery and downy mildew infection. BMC Plant Biol. 2021, 21, 265. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, G.; Upadhyay, S.K.; Singh, K. Vitis vinifera (Grapevine) lncRNAs are potential regulators of response to necrotrophic fungus, Botrytis cinerea infection. Physiol. Mol. Plant Pathol. 2020, 112, 101553. [Google Scholar] [CrossRef]
- Liu, W.; Liu, J.; Ning, Y.; Ding, B.; Wang, X.; Wang, Z.; Wang, G.L. Recent progress in understanding PAMP- and effector-triggered immunity against the rice blast fungus Magnaporthe oryzae. Mol. Plant 2013, 6, 605–620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dodds, P.N.; Rathjen, J.P. Plant immunity: Towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 2010, 11, 539–548. [Google Scholar] [CrossRef]
- Maekawa, T.; Kufer, T.A.; Schulze-Lefert, P. NLR functions in plant and animal immune systems: So far and yet so close. Nat. Immunol. 2011, 12, 817–826. [Google Scholar] [CrossRef]
- Gassmann, W.; Bhattacharjee, S. Effector-triggered immunity signaling: From gene-for-gene pathways to protein– protein interaction networks. Mol. Plant–Microbe Interact. 2012, 25, 862–868. [Google Scholar] [CrossRef] [Green Version]
- Boller, T.; Felix, G. A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 2009, 60, 379–406. [Google Scholar] [CrossRef]
- de Jonge, R.; van Esse, H.P.; Maruthachalam, K.; Bolton, M.D.; Santhanam, P.; Saber, M.K.; Zhang, Z.; Usami, T.; Lievens, B.; Subbarao, K.V.; et al. Tomato immune receptor Ve1 recognizes effector of multiple fungal pathogens uncovered by genome and RNA sequencing. Proc. Natl Acad. Sci. USA 2012, 109, 5110–5115. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.; Li, J.; Zhang, X.; Zhang, Q.; Huang, J.; Chen, J.Q.; Hartl, D.L.; Tian, D. Rapidly evolving R genes in diverse grass species confer resistance to rice blast disease. Proc. Natl Acad. Sci. USA 2013, 110, 18572–18577. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Liu, J.; Triplett, L.; Leach, J.E.; Wang, G.L. Novel insights into rice innate immunity against bacterial and fungal pathogens. Annu. Rev. Phytopathol. 2014, 52, 213–241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, L.; Chen, Y.; Luo, L.; Peck, S.C. Central roles and regulatory mechanisms of dual-specificity MAPK phosphatases in developmental and stress signaling. Front. Plant Sci. 2018, 9, 1697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Liu, T.; Shen, D.; Wang, J.; Ling, X.; Hu, Z.; Chen, T.; Hu, J.; Huang, J.; Yu, W.; et al. Tomato yellow leaf curl virus intergenic siRNAs target a host long noncoding RNA to modulate disease symptoms. PLoS Pathog. 2019, 15, e1007534. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Liang, Q.; Li, C.; Fu, S.; Kundu, J.K.; Zhou, X.; Wu, J. Transcriptome Analysis of Rice Reveals the lncRNA-mRNA Regulatory Network in Response to Rice Black-Streaked Dwarf Virus Infection. Viruses 2020, 12, 951. [Google Scholar] [CrossRef]
- Van Loon, L.C.; Rep, M.; Pieterse, C.M. Significance of inducible defense-related proteins in infected plants. Annu. Rev.Phytopathol. 2006, 44, 135–162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bednarek, P. Chemical warfare or modulators of defence responses—The function of secondary metabolites in plant immunity. Curr. Opin. Plant Biol. 2012, 15, 407–414. [Google Scholar] [CrossRef] [PubMed]



| Database | Features | Reference |
|---|---|---|
| Plant long non-coding RNA database | This database consists of >13,000 lincRNAs and associated epigenetic markers | [18] |
| Plant ncRNA database | It consists of 11 different types of ncRNAs of 150 plant species | [30] |
| Green non-coding database | It consists of data of 37 plant species and algae with more than 120,000 lncRNAs | [31] |
| The Arabidopsis information resource | It also consists of data of various noncoding RNAs | [32] |
| Araportll | It consists of annotated lincRNA, NATs, and various other ncRNAs | [33] |
| Plant natural antisense transcripts database | It consists of NATs annotated data along with expression of small RNA of 70 plant species | [34] |
| CANTATAdb | It consists of data of 45,000 lncRNAs of 10 model plant species | [35] |
| Pathogen | Associated Stress | lncRNA | Mechanism | Plant | Reference |
|---|---|---|---|---|---|
| Bacteria | Bacterial speck disease (Pseudomonas syringae pv. tomato DC3000) | Up- ELENA1 | Directly interact with MED19a | Arabidopsis thaliana | [55] |
| Bacterial canker (Pseudomonas syringae pv. actinidiae) | Up- TCONS_00202033, TCONS_0019494 & TCONS_00076221 | Unknown | Actinidi adeliciosa | [56] | |
| Bacterial leaf blight (Xanthomonas oryzae pv. oryzae) | Up- ALEX1 | Interacts with JA related genes | Oryza sativa | [57] | |
| Fungal | Powdery mildew (Blumeria graminis f. sp. tritici) | Up- TalnRNA5, TapmlnRNA19 | Precursor of miR2004 | Triticum aestivum | [58] |
| Powdery mildew (Blumeria graminis f. sp. tritici) | Up- TalnRNA9 | Signal recognition particle 7S RNA variant 1 | Triticum aestivum | [58] | |
| Powdery mildew (Blumeria graminis f. sp. tritici) | Up- TapmlnRNA2, TapmlnRNA7 | Precursor of siRNA | Triticum aestivum | [58] | |
| White mold (Sclerotinia sclerotiorum) | Up- TCONS_00012499, TCONS_00004577, TCONS_00004034, TCONS_00009614, TCONS_00015411 | Precursor of mi156 | Brassica napus | [59] | |
| White mold (Sclerotinia sclerotiorum) | Up- TCONS_00006568, TCONS_00018692, TCONS_000017152, TCONS_00008591, TCONS_00001092 | Precursor of mi169 | Brassica napus | [59] | |
| Wilt disease (Fusarium oxysporum) | Up- TAR-66 (lincRNA) | Co-induction with neighboring defense-related gene | Arabidopsis thaliana | [60] | |
| Wilt disease (Fusarium oxysporum) | Up- TAR- 67,-191,- 197,-224 | Unknown | Arabidopsis thaliana | [60] | |
| Stripe rust (Puccinia striiformis f. sp. tritici) | Up- TalncRNA18, 106 | Unknown | Triticum aestivum | [61] | |
| Stripe rust (Puccinia striiformis f. sp. tritici) | Up & Dp at different dpi- TalncRNA73, 108 | Unknown | Triticum aestivum | [61] | |
| Viral | TYLCV Infection | Up- Slylnc0195 | Target mimicry of miR166 | Solanum lycopersicum | [62] |
| TYLCVInfection | Dn- Slylnc1077 | Target mimicry of miR399 | Solanum lycopersicum | [62] | |
| CGMMV infection | Up- lncRNALNC_1497 | Target mimicry of MIR4995-p5_Iss19GC | Citrullus lanatus | [63] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sharma, Y.; Sharma, A.; Madhu; Shumayla; Singh, K.; Upadhyay, S.K. Long Non-Coding RNAs as Emerging Regulators of Pathogen Response in Plants. Non-Coding RNA 2022, 8, 4. https://0-doi-org.brum.beds.ac.uk/10.3390/ncrna8010004
Sharma Y, Sharma A, Madhu, Shumayla, Singh K, Upadhyay SK. Long Non-Coding RNAs as Emerging Regulators of Pathogen Response in Plants. Non-Coding RNA. 2022; 8(1):4. https://0-doi-org.brum.beds.ac.uk/10.3390/ncrna8010004
Chicago/Turabian StyleSharma, Yashraaj, Alok Sharma, Madhu, Shumayla, Kashmir Singh, and Santosh Kumar Upadhyay. 2022. "Long Non-Coding RNAs as Emerging Regulators of Pathogen Response in Plants" Non-Coding RNA 8, no. 1: 4. https://0-doi-org.brum.beds.ac.uk/10.3390/ncrna8010004