Delineating Management Zones with Different Yield Potentials in Soybean–Corn and Soybean–Cotton Production Systems
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Plots
2.2. Data Sets
2.3. Vegetation Indices Data Sets
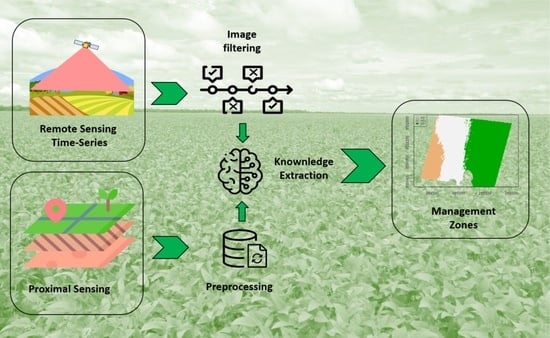
2.4. Delineation of Management Zones
2.5. Yield Variance Reduction
2.6. Attribute Selection Procedures for MZ Delineation
2.7. Methodology Summary
3. Results
3.1. Procedure 1: MZs Using Field Data and Peak-Biomass VIs
3.2. Procedure 2: MZs Using Well-Correlated VIs and Soil Attributes
3.3. Procedure 3: MZs Using VIs Replacing Field Data
3.4. Procedure 4: MZs Using VIs Selected by Variable Importance Model
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Moral, F.J.; Terrón, J.M.; Da Silva, J.M. Delineation of management zones using mobile measurements of soil apparent electrical conductivity and multivariate geostatistical techniques. Soil Tillage Res. 2010, 106, 335–343. [Google Scholar] [CrossRef]
- Damian, J.M.; Pias, O.H.D.C.; Cherubin, M.R.; Fonseca, A.Z.D.; Fornari, E.Z.; Santi, A.L. Applying the NDVI from satellite images in delimiting management zones for annual crops. Sci. Agric. 2019, 77, e20180055. [Google Scholar] [CrossRef]
- Ali, A.; Rondelli, V.; Martelli, R.; Falsone, G.; Lupia, F.; Barbanti, L. Management zones delineation through clustering techniques based on soils traits, NDVI data, and multiple year crop yields. Agriculture 2022, 12, 231. [Google Scholar] [CrossRef]
- Adiele, J.G.; Schut, A.G.; Van Den Beuken, R.P.M.; Ezui, K.S.; Pypers, P.; Ano, A.O.; Giller, K.E. Towards closing cassava yield gap in West Africa: Agronomic efficiency and storage root yield responses to NPK fertilizers. Field Crops Res. 2020, 253, 107820. [Google Scholar] [CrossRef]
- Moura, S.S.; Franca, L.T.; Pereira, V.S.; Teodoro, P.E.; Baio, F.H. Seeding rate in soybean according to the soil apparent electrical conductivity. An. Acad. Bras. Ciências 2020, 92, 107820. [Google Scholar] [CrossRef]
- García-Martínez, H.; Flores-Magdaleno, H.; Ascencio-Hernández, R.; Khalil-Gardezi, A.; Tijerina-Chávez, L.; Mancilla-Villa, O.R.; Vázquez-Peña, M.A. Corn grain yield estimation from vegetation indices, canopy cover, plant density, and a neural network using multispectral and RGB images acquired with unmanned aerial vehicles. Agriculture 2020, 10, 277. [Google Scholar] [CrossRef]
- Vasconcelos, J.C.S.; Speranza, E.A.; Antunes, J.F.G.; Barbosa, L.A.F.; Christofoletti, D.; Severino, F.J.; de Almeida Cançado, G.M. Development and Validation of a Model Based on Vegetation Indices for the Prediction of Sugarcane Yield. AgriEngineering 2023, 5, 698–719. [Google Scholar] [CrossRef]
- Gavioli, A.; de Souza, E.G.; Bazzi, C.L.; Schenatto, K.; Betzek, N.M. Identification of management zones in precision agriculture: An evaluation of alternative cluster analysis methods. Biosyst. Eng. 2019, 181, 86–102. [Google Scholar] [CrossRef]
- Bottega, E.L.; de Queiroz, D.M.; de Assis de Carvalho Pinto, F.; de Souza, C.M.A.; Valente, D.S.M. Precision agriculture applied to soybean: Part I-Delineation of management zones. Aust. J. Crop Sci. 2017, 11, 573–579. [Google Scholar] [CrossRef]
- Santos, R.T.; Saraiva, A.M. A reference process for management zones delineation in precision agriculture. IEEE Lat. Am. Trans. 2015, 13, 727–738. [Google Scholar] [CrossRef]
- Reyes, J.; Wendroth, O.; Matocha, C.; Zhu, J. Delineating site-specific management zones and evaluating soil water temporal dynamics in a farmer’s field in Kentucky. Vadose Zone J. 2019, 18, 1–19. [Google Scholar] [CrossRef]
- Georgi, C.; Spengler, D.; Itzerott, S.; Kleinschmit, B. Automatic delineation algorithm for site-specific management zones based on satellite remote sensing data. Precis. Agric. 2018, 19, 684–707. [Google Scholar] [CrossRef]
- Leo, S.; Migliorati, M.A.; Nguyen, T.H.; Grace, P.R. Combining remote sensing-derived management zones and an auto-aclibrated crop simulation model to determine optimal nitrogen fertilizer rates. Agric. Syst. 2023, 205, 103559. [Google Scholar] [CrossRef]
- Maia, F.C.O.; Bufon, V.B.; Leão, T.P. Vegetation indices as a Tool for Mapping Sugarcane Managemenz Zones. Precis. Agric. 2023, 24, 213–234. [Google Scholar] [CrossRef]
- Huete, A.; Didan, K.; Miura, T.; Rodriguez, E.P.; Gao, X.; Ferreira, L.G. Overview of the radiometric and biophysical performance of the MODIS vegetation indices. Remote Sens. Environ. 2002, 83, 195–213. [Google Scholar] [CrossRef]
- Rouse, J.W.; Haas, R.H.; Schell, J.A.; Deering, D.W. Monitoring vegetation systems in the Great Plains with ERTS. NASA Spec. Publ. 1974, 351, 309. [Google Scholar]
- Rahul, T.; Kumar, N.A.; Biswaranjan, D.; Mohammad, S.; Banwari, L.; Priyanka, G.; Kumar, S.A. Assessing soil spatial variability and delineating site-specific management zones for a coastal saline land in eastern India. Arch. Agron. Soil Sci. 2019, 65, 1775–1787. [Google Scholar] [CrossRef]
- Pearson, R.L.; Miller, L.D. Remote mapping of standing crop biomass for estimation of the productivity of the shortgrass prairie. Remote Sens. Environ. 1972, VIII, 1355. [Google Scholar]
- Merzlyak, M.N.; Gitelson, A.A.; Chivkunova, O.B.; Rakitin, V.Y. Non-destructive optical detection of pigment changes during leaf senescence and fruit ripening. Physiol. Plant. 1999, 106, 135–141. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Kaufman, Y.J.; Merzlyak, M.N. Use of a green channel in remote sensing of global vegetation from EOS-MODIS. Remote Sens. Environ. 1996, 58, 289–298. [Google Scholar] [CrossRef]
- Broge, N.H.; Leblanc, E. Comparing prediction power and stability of broadband and hyperspectral vegetation indices for estimation of green leaf area index and canopy chlorophyll density. Remote Sens. Environ. 2001, 76, 156–172. [Google Scholar] [CrossRef]
- Vincini, M.; Frazzi, E.R.M.E.S.; D’Alessio, P.A.O.L.O. A broad-band leaf chlorophyll vegetation index at the canopy scale. Precis. Agric. 2008, 9, 303–319. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Gritz, Y.; Merzlyak, M.N. Relationships between leaf chlorophyll content and spectral reflectance and algorithms for non-destructive chlorophyll assessment in higher plant leaves. J. Plant Physiol. 2003, 160, 271–282. [Google Scholar] [CrossRef]
- Jordan, C.F. Derivation of leaf-area index from quality of light on the forest floor. Ecology 1969, 50, 663–666. [Google Scholar] [CrossRef]
- Gitelson, A.; Merzlyak, M.N. Spectral reflectance changes associated with autumn senescence of Aesculus hippocastanum L. and Acer platanoides L. leaves. Spectral features and relation to chlorophyll estimation. J. Plant Physiol. 1994, 143, 286–292. [Google Scholar] [CrossRef]
- Huete, A.R. A soil-adjusted vegetation index (SAVI). Remote Sens. Environ. 1988, 25, 295–309. [Google Scholar] [CrossRef]
- Pelleg, D.; Moore, A.W. X-means: Extending k-means with efficient estimation of the number of clusters. ICML 2000, 1, 727–734. [Google Scholar]
- Maldaner, L.F.; de Paula Corrêdo, L.; Canata, T.F.; Molin, J.P. Predicting the sugarcane yield in real-time by harvester engine parameters and machine learning approaches. Comput. Electron. Agric. 2021, 181, 105945. [Google Scholar] [CrossRef]
- Dobermann, A.; Ping, J.L.; Adamchuk, V.I.; Simbahan, G.C.; Ferguson, R.B. Classification of crop yield variability in irrigated production fields. Agron. J. 2003, 95, 1105–1120. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]



| Plot A | |||||
|---|---|---|---|---|---|
| Attribute | Source * | Samples/ha | Spatial Int. ** | Mean *** | SD **** |
| Clay (%) | Soil Sampling | 1 | Kriging | 47.6 | 18.7 |
| Elevation (m) | Harvest GPS | 785 | IDW | 555.5 | 9.6 |
| Cotton Yield-2019 (ton/ha) | Harvest Monitor | 785 | IDW | 3.94 | 0.80 |
| Cotton Yield-2020 (ton/ha) | Harvest Monitor | 595 | IDW | 4.23 | 0.70 |
| Soybean Yield-2021 (ton/ha) | Harvest Monitor | 20 | IDW | 3.76 | 0.32 |
| Soybean Yield-2022 (ton/ha) | Harvest Monitor | 28 | IDW | 0.97 | 0.25 |
| Plot B | |||||
| Attribute | Source * | Samples/ha | Spatial Int. ** | Mean *** | SD **** |
| Clay (%) | Soil Sampling | 1 | Kriging | 39.5 | 11.9 |
| Elevation (m) | Harvest GPS | 762 | IDW | 558.9 | 6.9 |
| Cotton Yield-2019 (ton/ha) | Harvest Monitor | 762 | IDW | 4.62 | 0.91 |
| Cotton Yield-2020 (ton/ha) | Harvest Monitor | 714 | IDW | 5.51 | 1.57 |
| Soybean Yield-2021(ton/ha) | Harvest Monitor | 14 | IDW | 3.38 | 0.79 |
| Soybean Yield-2022 (ton/ha) | Harvest Monitor | 34 | IDW | 3.05 | 1.12 |
| Plot C | |||||
| Attribute | Source * | Samples/ha | Spatial Int. ** | Mean *** | SD **** |
| Clay (%) | Soil Sampling | 5 | Kriging | 45.9 | 1.30 |
| Potassium (x) | Soil Sampling | 5 | Kriging | 0.14 | 0.03 |
| Cotton Yield-2022 (ton/ha) | Harvest Monitor | 530 | IDW | 6.98 | 1.91 |
| Plot D | |||||
| Attribute | Source * | Samples/ha | Spatial Int. ** | Mean *** | SD **** |
| ECa (0–50 cm) | SoilXplorer Sensor | 326 | Kriging | 61.5 | 9.49 |
| Soybean Yield (2022) (ton/ha) | Harvest Monitor | 1265 | Kriging | 4.0 | 0.43 |
| Corn Yield (2022) (ton/ha) | Harvest Monitor | 1265 | Kriging | 7.6 | 1.15 |
| Elevation (m) | GPS Combine Harvest Monitor | 1265 | Kriging | 632 | 16 |
| Acronym | Name | Formula * | Reference |
|---|---|---|---|
| NDVI | Normalized Difference Vegetation Index | (NIR − R)/(NIR+R) | [16] |
| RVI | Ratio Vegetation Index | NIR/R | [18] |
| PSRI | Plant Senescence Reflectance Index | (R − G)/RE | [19] |
| GNDVI | Green Normalized Difference Vegetation Index | (NIR − G)/(NIR+G) | [20] |
| TVI | Triangular Vegetation Index | 0.5 × (120 × (NIR − G) − 200 × (R − G)) | [21] |
| CVI | Chlorophyll Vegetation Index | (NIR × R)/(G2) | [22] |
| CIG | Chlorophyll Index—Green | (NIR/G) − 1 | [23] |
| CIRE | Chlorophyll Index—Red Edge | (NIR/RE) − 1 | [23] |
| DVI | Difference Vegetation Index | NIR-RE | [24] |
| NDRE | Normalized Difference Red Edge Index | (NIR-RE)/(NIR+RE) | [25] |
| EVI | Enhanced Vegetation Index | 2.5 × (NIR − R)/(NIR+6 × R − 7.5 × B+1) | [15] |
| SAVI | Soil-Adjusted Vegetation Index | (NIR − R)/(NIR+R+0.428) × (1.428) | [26] |
| VI | Plot A | Plot B | Plot C | Plot D |
|---|---|---|---|---|
| NDVI | 0.553 | 0.585 | 0.451 | 0.469 |
| RVI | 0.526 | 0.149 | 0.425 | 0.346 |
| PSRI | 0.294 | 0.196 | 0.438 | 0.030 |
| GNDVI | 0.640 | 0.616 | 0.518 | 0.418 |
| TVI | 0.659 | 0.641 | 0.533 | 0.527 |
| CVI | 0.659 | 0.641 | −0.125 | 0.527 |
| CIG | 0.660 | 0.637 | 0.533 | 0.528 |
| CIRE | 0.302 | 0.225 | 0.313 | 0.231 |
| DVI | 0.302 | 0.255 | 0.313 | 0.231 |
| NDRE | 0.318 | 0.353 | 0.030 | 0.053 |
| EVI | 0.668 | 0.644 | 0.521 | 0.525 |
| SAVI | 0.587 | 0.611 | 0.534 | 0.485 |
| Plot | Attributes | MZ Map | * Av. Yield Map | VR (%) |
|---|---|---|---|---|
| A | Clay * Av. Yield * Av. EVI |  |  | 70.1 |
| B | Clay * Av. Yield * Av. EVI |  |  | 61.5 |
| C | Clay Potassium *Av. Yield EVI |  |  | 0.1 |
| D | ECa Elevation *Av. Yield NDVI-Winter NDVI-Summer |  |  | 35.2 |
| Plot | Attributes | MZ Map | * Av. Yield Map | VR (%) |
|---|---|---|---|---|
| A | Clay EVI CVI |  |  | 64.0 |
| B | Clay EVI CVI |  |  | 50.7 |
| C | Clay EVI SAVI CIG TVI |  |  | 1.3 |
| D | ECa CVI EVI CIG TVI |  |  | 15.4 |
| Plot | Attributes | MZ Map | * Av. Yield Map | VR (%) |
|---|---|---|---|---|
| A | GNDVI |  |  | 40.3 |
| B | GNDVI CIG |  |  | 41.6 |
| C | TVI EVI SAVI CIG |  |  | 1.3 |
| Plot | Attributes | MZ Map | * Av. Yield Map | VR (%) |
|---|---|---|---|---|
| A | SAVI PSRI NDRE GNDVI |  |  | 47.9 |
| B | PSRI NDRE GNDVI |  |  | 42.3 |
| C | PSRI GNDVI SAVI |  |  | 0.84 |
| D | PSRI SAVI |  |  | 4.0 |
| Plot A | Plot B | Plot C | Plot D | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Procedure | MZ1 | MZ2 | MZ3 | MZ4 | MZ1 | MZ2 | MZ3 | MZ4 | MZ1 | MZ2 | MZ3 | MZ4 | MZ1 | MZ2 |
| 1 | 0 | −0.2 | 0.1 | - | −0.2 | 0.0 | - | - | −0.008 | 0.003 | - | - | 0.2 | −0.2 |
| 2 | 0 | −0.2 | 0.1 | - | 0.0 | −0.2 | 0.1 | 0.0 | 0.02 | 0.0 | −0.04 | 0.2 | −01 | |
| 3 | −0.2 | 0 | - | - | 0.0 | −0.2 | - | - | 0.0 | 0.01 | −0.02 | −0.07 | - | - |
| 4 | −0.1 | −0.2 | 0.0 | 0.1 | 0.0 | −0.2 | - | - | 0.0 | −0.03 | - | - | 0.05 | −0.06 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Speranza, E.A.; Naime, J.d.M.; Vaz, C.M.P.; Santos, J.C.F.d.; Inamasu, R.Y.; Lopes, I.d.O.N.; Queirós, L.R.; Rabelo, L.M.; Jorge, L.A.d.C.; Chagas, S.d.; et al. Delineating Management Zones with Different Yield Potentials in Soybean–Corn and Soybean–Cotton Production Systems. AgriEngineering 2023, 5, 1481-1497. https://0-doi-org.brum.beds.ac.uk/10.3390/agriengineering5030092
Speranza EA, Naime JdM, Vaz CMP, Santos JCFd, Inamasu RY, Lopes IdON, Queirós LR, Rabelo LM, Jorge LAdC, Chagas Sd, et al. Delineating Management Zones with Different Yield Potentials in Soybean–Corn and Soybean–Cotton Production Systems. AgriEngineering. 2023; 5(3):1481-1497. https://0-doi-org.brum.beds.ac.uk/10.3390/agriengineering5030092
Chicago/Turabian StyleSperanza, Eduardo Antonio, João de Mendonça Naime, Carlos Manoel Pedro Vaz, Júlio Cezar Franchini dos Santos, Ricardo Yassushi Inamasu, Ivani de Oliveira Negrão Lopes, Leonardo Ribeiro Queirós, Ladislau Marcelino Rabelo, Lucio André de Castro Jorge, Sergio das Chagas, and et al. 2023. "Delineating Management Zones with Different Yield Potentials in Soybean–Corn and Soybean–Cotton Production Systems" AgriEngineering 5, no. 3: 1481-1497. https://0-doi-org.brum.beds.ac.uk/10.3390/agriengineering5030092