The Clinical and Biological Effects of PD-1 Expression on Tumor Cells in Diffuse Large B-Cell Lymphoma
Abstract
:1. Introduction
2. Results
2.1. Patient Characteristics
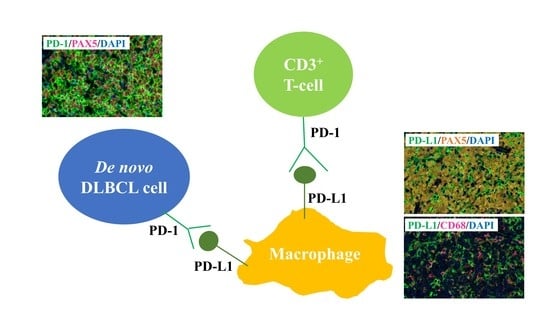
2.2. PD-1 and PD-L1 Expression Detected Using MC-IF
2.3. Correlation between PD-1 Expression on Lymphoma Cells and Clinical Relevance
2.4. Biological Effects of PD-1 KO via CRISPR-Cas9-Mediated Genome Editing in Human B-Lymphoma VAL Cells
3. Discussion
4. Materials and Methods
4.1. Tissue Samples and Antibodies Used for MC-IF Staining
4.2. MC-IF Analysis
4.3. Cell Culture and Reagents
4.4. PD-1 Knockout Using the Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)-Cas9 System
4.5. Western Blotting and Flow Cytometry
4.6. Microarray Gene Expression Analyses for VAL/Mock and VAL/PD-1-KO Cells
4.7. Cell Viability Assay
4.8. Soft Agar Colony Formation Assay
4.9. Cell Migration Assay
4.10. Annexin V Assay
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Iwai, Y.; Okazaki, T.; Nishimura, H.; Kawasaki, A.; Yagita, H.; Honjo, T. Microanatomical localization of PD-1 in human tonsils. Immunol. Lett. 2002, 83, 215–220. [Google Scholar] [CrossRef]
- Salmaninejad, A.; Valilou, S.F.; Shabgah, A.G.; Aslani, S.; Alimardani, M.; Pasdar, A.; Sahebkar, A. PD-1/PD-L1 pathway: Basic biology and role in cancer immunotherapy. J. Cell. Physiol. 2019, 234, 16824–16837. [Google Scholar] [CrossRef]
- Du, S.; McCall, N.; Park, K.; Guan, Q.; Fontina, P.; Ertel, A.; Zhan, T.; Dicker, A.P.; Lu, B. Blockade of Tumor-Expressed PD-1 promotes lung cancer growth. OncoImmunology 2018, 7, e1408747. [Google Scholar] [CrossRef] [PubMed]
- Kleffel, S.; Posch, C.; Barthel, S.R.; Mueller, H.; Schlapbach, C.; Guenova, E.; Elco, C.P.; Lee, N.; Juneja, V.R.; Zhan, Q.; et al. Melanoma cell-Intrinsic PD-1 receptor functions promote tumor growth. Cell 2015, 162, 1242–1256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kiyasu, J.; Miyoshi, H.; Hirata, A.; Arakawa, F.; Ichikawa, A.; Niino, D.; Sugita, Y.; Yufu, Y.; Choi, I.; Abe, Y.; et al. Expression of programmed cell death ligand 1 is associated with poor overall survival in patients with diffuse large B-cell lymphoma. Blood 2015, 126, 2193–2201. [Google Scholar] [CrossRef] [PubMed]
- Xu-Monette, Z.Y.; Zhou, J.; Young, K.H. PD-1 expression and clinical PD-1 blockade in B-cell lymphomas. Blood 2018, 131, 68–83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gravelle, P.; Burroni, B.; Pericart, S.; Rossi, C.; Bezombes, C.; Tosolini, M.; Damotte, D.; Brousset, P.; Fournié, J.J.; Laurent, C. Mechanisms of PD-1/PD-L1 expression and prognostic relevance in non-Hodgkin lymphoma: A summary of immunohistochemical studies. Oncotarget 2017, 8, 44960–44975. [Google Scholar] [CrossRef] [Green Version]
- Xu-Monette, Z.Y.; Xiao, M.; Au, Q.; Padmanabhan, R.; Xu, B.; Hoe, N.; Rodríguez-Perales, S.; Torres-Ruiz, R.; Manyam, G.C.; Visco, C.; et al. Immune profiling and quantitative analysis decipher the clinical role of immune-checkpoint expression in the tumor immune microenvironment of DLBCL. Cancer Immunol. Res. 2019, 7, 644–657. [Google Scholar] [CrossRef]
- Ansell, S.M.; Minnema, M.C.; Johnson, P.; Timmerman, J.M.; Armand, P.; Shipp, M.A.; Rodig, S.J.; Ligon, A.H.; Roemer, M.G.M.; Reddy, N.; et al. Nivolumab for relapsed/refractory diffuse large B-cell lymphoma in patients ineligible for or having failed autologous transplantation: A single-arm, phase II study. J. Clin. Oncol. 2019, 37, 481–489. [Google Scholar] [CrossRef]
- Iwai, Y.; Hamanishi, J.; Chamoto, K.; Honjo, T. Cancer immunotherapies targeting the PD-1 signaling pathway. J. Biomed. Sci. 2017, 24, 26. [Google Scholar] [CrossRef] [Green Version]
- Champiat, S.; Dercle, L.; Ammari, S.; Massard, C.; Hollebecque, A.; Postel-Vinay, S.; Chaput, N.; Eggermont, A.; Marabelle, A.; Soria, J.C.; et al. Hyperprogressive disease is a new pattern of progression in cancer patients treated by anti-PD-1/PD-L1. Clinical. Cancer Res. 2017, 23, 1920–1928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ratner, L.; Waldmann, T.A.; Janakiram, M.; Brammer, J.E. Rapid progression of adult T-cell leukemia–lymphoma after PD-1 inhibitor therapy. N. Engl. J. Med. 2018, 378, 1947–1948. [Google Scholar] [CrossRef] [PubMed]
- Stack, E.C.; Wang, C.; Roman, K.A.; Hoyt, C.C. Multiplexed immunohistochemistry, imaging, and quantitation: A review, with an assessment of Tyramide signal amplification, multispectral imaging and multiplex analysis. Methods 2014, 70, 46–58. [Google Scholar] [CrossRef]
- Feng, Z.; Puri, S.; Moudgil, T.; Wood, W.; Hoyt, C.C.; Wang, C.; Urba, W.J.; Curti, B.D.; Bifulco, C.B.; Fox, B.A. Multispectral imaging of formalin-fixed tissue predicts ability to generate tumor-infiltrating lymphocytes from melanoma. J. Immunother. Cancer 2015, 3, 47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carey, C.D.; Gusenleitner, D.; Lipschitz, M.; Roemer, M.G.M.; Stack, E.C.; Gjini, E.; Hu, X.; Redd, R.; Freeman, G.J.; Neuberg, D.; et al. Topological analysis reveals a PD-L1-associated microenvironmental niche for Reed-Sternberg cells in Hodgkin lymphoma. Blood 2017, 130, 2420–2430. [Google Scholar] [CrossRef] [PubMed]
- Muenst, S.; Hoeller, S.; Willi, N.; Dirnhofera, S.; Tzankov, A. Diagnostic and prognostic utility of PD-1 in B cell lymphomas. Dis. Markers 2010, 29, 47–53. [Google Scholar] [CrossRef]
- Laurent, C.; Charmpi, K.; Gravelle, P.; Tosolini, M.; Franchet, C.; Ysebaert, L.; Brousset, P.; Bidaut, A.; Ycart, B.; Fournié, J.J. Several immune escape patterns in non-Hodgkin’s lymphomas. Oncoimmunology 2015, 4, e1026530. [Google Scholar] [CrossRef] [Green Version]
- McCord, R.; Bolen, C.R.; Koeppen, H.; Kadel, E.E., 3rd; Oestergaard, M.Z.; Nielsen, T.; Sehn, L.H.; Venstrom, J.M. PD-L1 and tumor-associated macrophages in de novo DLBCL. Blood Adv. 2019, 3, 531–540. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Heazlewood, S.Y.; Williams, B.; Cardozo, D.; Nigro, J.; Oteiza, A.; Nilsson, S.K. The role of CD44 in fetal and adult hematopoietic stem cell regulation. Haematologica 2016, 101, 26–37. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Xiao, Z.; Wong, S.K.; Tin, V.P.; Ho, K.Y.; Wang, J.; Sham, M.H.; Wong, M.P. Lung cancer tumorigenicity and drug resistance are maintained through ALDH(hi)CD44(hi) tumor initiating cells. Oncotarget 2013, 4, 1698–1711. [Google Scholar] [CrossRef] [Green Version]
- Schatton, T.; Schütte, U.; Frank, N.Y.; Zhan, Q.; Hoerning, A.; Robles, S.C.; Zhou, J.; Hodi, F.S.; Spagnoli, G.C.; Murphy, G.F.; et al. Modulation of T-cell activation by malignant melanoma initiating cells. Cancer Res. 2010, 70, 697–708. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.W.; Liu, L.; Gong, C.Y.; Shi, H.S.; Zeng, Y.H.; Wang, X.Z.; Zhao, Y.W.; Wei, Y.Q. Prognostic significance of tumor-associated macrophages in solid tumor: A meta-analysis of the literature. PLoS ONE 2012, 7, e50946. [Google Scholar] [CrossRef] [Green Version]
- Gwak, J.M.; Jang, M.H.; Kim, D.I.; Seo, A.N.; Park, S.Y. Prognostic value of tumor-associated macrophages according to histologic locations and hormone receptor status in breast cancer. PLoS ONE 2015, 10, e0125728. [Google Scholar] [CrossRef]
- Cassetta, L.; Pollard, J.W. Targeting macrophages: Therapeutic approaches in cancer. Nat. Rev. Drug. Discov. 2018, 17, 887–904. [Google Scholar] [CrossRef] [PubMed]
- Rossille, D.; Gressier, M.; Damotte, D.; Maucort-Boulch, D.; Pangault, C.; Semana, G.; Le Gouill, S.; Haioun, C.; Tarte, K.; Lamy, T.; et al. Groupe Ouest-Est des Leucémies et Autres Maladies du Sang; Groupe Ouest-Est des Leucémies et Autres Maladies du Sang. High level of soluble programmed cell death ligand 1 in blood impacts overall survival in aggressive diffuse large B-Cell lymphoma: Results from a French multicenter clinical trial. Leukemia 2014, 28, 2367–2375. [Google Scholar] [PubMed]
- Thibult, M.L.; Mamessier, E.; Gertner-Dardenne, J.; Pastor, S.; Just-Landi, S.; Xerri, L.; Chetaille, B.; Olive, D. PD-1 is a novel regulator of human B-cell activation. Int. Immunol. 2012, 25, 129–137. [Google Scholar] [CrossRef] [Green Version]
- Juweid, M.E.; Wiseman, G.A.; Vose, J.M.; Ritchie, J.M.; Menda, Y.; Wooldridge, J.E.; Mottaghy, F.M.; Rohren, E.M.; Blumstein, N.M.; Stolpen, A.; et al. Response assessment of aggressive non-Hodgkin’s lymphoma by integrated International Workshop Criteria and fluorine-18–fluorodeoxyglucose positron emission tomography. J. Clin. Oncol. 2005, 23, 4652–4661. [Google Scholar] [CrossRef] [PubMed]
- Cheson, B.D.; Pfistner, B.; Juweid, M.E.; Gascoyne, R.D.; Specht, L.; Horning, S.J.; Coiffier, B.; Fisher, R.I.; Hagenbeek, A.; Zucca, E.; et al. International Harmonization Project on Lymphoma. Revised response criteria for malignant lymphoma. J. Clin. Oncol. 2007, 25, 579–586. [Google Scholar] [CrossRef]
- Wahiduzzaman, M.; Ota, A.; Karnan, S.; Hanamura, I.; Mizuno, S.; Kanasugi, J.; Rahman, M.L.; Hyodo, T.; Konishi, H.; Tsuzuki, S.; et al. Novel combined Ato-C treatment synergistically suppresses proliferation of Bcr-Abl-positive leukemic cells in vitro and in vivo. Cancer Lett. 2018, 433, 117–130. [Google Scholar] [CrossRef] [PubMed]
- Asai, A.; Karnan, S.; Ota, A.; Takahashi, M.; Damdindorj, L.; Konishi, Y.; Hossain, E.; Konishi, H.; Nagata, A.; Yokoo, K.; et al. High-resolution 400K oligonucleotide array comparative genomic hybridization analysis of neurofibromatosis type 1-associated cutaneous neurofibromas. Gene 2015, 558, 220–226. [Google Scholar] [CrossRef]
- Huber, W.; von Heydebreck, A.; Sültmann, H.; Poustka, A.; Vingron, M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 2002, 18, S96–S104. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mootha, V.K.; Lindgren, C.M.; Eriksson, K.F.; Subramanian, A.; Sihag, S.; Lehar, J.; Puigserver, P.; Carlsson, E.; Ridderstråle, M.; Laurila, E.; et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003, 34, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Kanda, Y. Investigation of the freely available easy-to-use software ’EZR’ for medical statistics. Bone. Marrow. Transplant. 2013, 48, 452–458. [Google Scholar] [CrossRef] [Green Version]





| Parameter | Number of Patients | |
|---|---|---|
| (n, %) | ||
| Age, years | ||
| Median | 73 (43–90) | |
| ≤60 | 5 | 16% |
| >60 | 27 | 84% |
| Sex | ||
| M | 20 | 63% |
| F | 12 | 37% |
| Serum LD | ||
| Normal | 14 | 44% |
| Elevated | 18 | 56% |
| Ann Arbor stage | ||
| I or II | 14 | 44% |
| III or IV | 18 | 56% |
| Bulky mass (>7 cm) | 3 | 9% |
| COO (by Hans) | ||
| GCB | 11 | 34% |
| Non-GC | 19 | 66% |
| N/A | 2 | |
| Primary treatments | ||
| R-CHOP or R-CHOP-like | 32 | 100% |
| Number of Patients | |||
|---|---|---|---|
| (n, %) | |||
| PAX5+ cells (B-lymphoma cells) | |||
| PD-1 | Positive | 19 | 59% |
| Negative | 13 | 41% | |
| N/A | 0 | ||
| PD-L1 | Positive | 1 | 3% |
| Negative | 29 | 97% | |
| N/A | 2 | ||
| CD68+ cells (macrophages) in TME | |||
| PD-L1 | Positive | 24 | 86% |
| Negative | 4 | 14% | |
| N/A | 4 | ||
| CD3+ cells (T-cells) in TME | |||
| PD-1 | Positive | 24 | 89% |
| Negative | 3 | 11% | |
| N/A | 5 | ||
| Parameter | Number of Patients | PD-1 Positive | p |
|---|---|---|---|
| (n = 32) | No/Total (%) | ||
| Age, years | |||
| ≤60 | 5 | 2/5 (40%) | 0.69 |
| >60 | 27 | 17/27 (63%) | |
| Serum LD | |||
| Normal | 14 | 6/14 (42%) | 0.55 |
| Elevated | 18 | 13/18 (72%) | |
| Ann Arbor stage | |||
| I or II | 14 | 7/14 (50%) | 0.77 |
| III or IV | 18 | 12/18 (66%) | |
| COO (by Hans) | |||
| GCB | 12 | 6/12 (50%) | 0.76 |
| Non-GCB | 18 | 13/18 (72%) | |
| N/A | 2 | 0/2 | |
| PD-L1 expression on tumor cells | |||
| Positive | 1 | 0/1 (0%) | N/A |
| Negative | 28 | 16/28 (57%) | |
| N/A | 3 | 3/3 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hanamura, I.; Suzuki, S.; Ota, A.; Murakami, S.; Satou, A.; Takahara, T.; Karnan, S.; Lam, V.Q.; Nakamura, A.; Takasugi, S.; et al. The Clinical and Biological Effects of PD-1 Expression on Tumor Cells in Diffuse Large B-Cell Lymphoma. Hemato 2021, 2, 368-382. https://0-doi-org.brum.beds.ac.uk/10.3390/hemato2020023
Hanamura I, Suzuki S, Ota A, Murakami S, Satou A, Takahara T, Karnan S, Lam VQ, Nakamura A, Takasugi S, et al. The Clinical and Biological Effects of PD-1 Expression on Tumor Cells in Diffuse Large B-Cell Lymphoma. Hemato. 2021; 2(2):368-382. https://0-doi-org.brum.beds.ac.uk/10.3390/hemato2020023
Chicago/Turabian StyleHanamura, Ichiro, Susumu Suzuki, Akinobu Ota, Satsuki Murakami, Akira Satou, Taishi Takahara, Sivasundaram Karnan, Vu Quang Lam, Ayano Nakamura, Souichi Takasugi, and et al. 2021. "The Clinical and Biological Effects of PD-1 Expression on Tumor Cells in Diffuse Large B-Cell Lymphoma" Hemato 2, no. 2: 368-382. https://0-doi-org.brum.beds.ac.uk/10.3390/hemato2020023