Changes in Ion Selectivity Following the Asymmetrical Addition of Charge to the Selectivity Filter of Bacterial Sodium Channels
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials Generation of Mutant Bacterial Channels
2.2. Cell Culture and Transfection
2.3. Electrophysiology
2.4. Equilibrium Simulations of NavMs Channel
2.5. Current-Voltage Curves Calculation
3. Results
3.1. Experimental Results
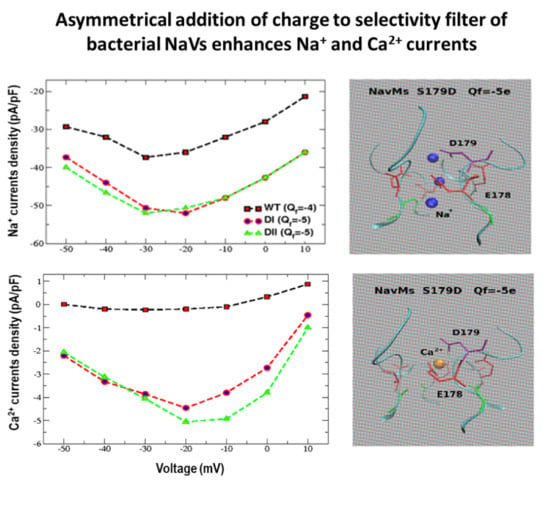
3.1.1. Na+/Ca2+ Selectivity for Randomly Mixed Populations of NaChBac Monomers
3.1.2. Na+/Ca2+ Selectivity for Concatenated NavMS Channels
3.2. Computational Results
4. Discussion and Conclusions
Supplementary Materials
Data Availability
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Catterall, W.A. Forty years of sodium channels: Structure, function, pharmacology, and epilepsy. Neurochem. Res. 2017, 42, 2495–2504. [Google Scholar] [CrossRef] [PubMed]
- Flucher, B.E. Skeletal muscle CaV1.1 channelopathies. Pflug. Arch. Eur. J. Physiool. 2020, 472, 739–754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Q.; Chen, J.; Qin, Y.; Wang, J.; Zhou, L. Mutations in voltage-gated L-type calcium channel: Implications in cardiac arrhythmia. Channels 2018, 12, 201–218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oyrer, J.; Maljevic, S.; Scheffer, I.E.; Berkovic, S.F.; Petrou, S.; Reid, C.A. Ion channels in genetic epilepsy: From genes and mechanisms to disease-targeted therapies. Pharmacol. Rev. 2018, 70, 142–173. [Google Scholar] [CrossRef] [PubMed]
- Catterall, W.A. Structure and regulation of voltage-gated Ca2+ channels. Annu. Rev. Cell. Dev. Biol. 2000, 16, 521–555. [Google Scholar] [CrossRef] [PubMed]
- Tyson, J.R.; Snutch, T.P. Molecular nature of voltage-gated calcium channels: Structure and species comparison. Wiley Interdiscrip. Rev. Membr. Transp. Signal. 2013, 2, 181–206. [Google Scholar] [CrossRef]
- Catterall, W.A.; Swanson, T.M. Structural basis for pharmacology of voltage-gated sodium and calcium channels. Mol. Pharmacol. 2015, 88, 141–150. [Google Scholar] [CrossRef]
- Zamponi, G.W.; Striessnig, J.; Koschak, A.; Dolphin, A.C. The physiology, pathology, and pharmacology of voltage-gated calcium channels and their future therapeutic potential. Pharmacol. Rev. 2015, 67, 821–870. [Google Scholar] [CrossRef] [Green Version]
- Xu, L.; Ding, X.; Wang, T.; Mou, S.; Sun, H.; Hou, T. Voltage-gated sodium channels: Structures, functions, and molecular modeling. Drug Discov. Today 2019, 24, 1389–1397. [Google Scholar] [CrossRef]
- Wu, J.; Yan, Z.; Li, Z.; Qian, X.; Lu, S.; Dong, M.; Zhou, Q.; Yan, M. Structure of the voltage-gated calcium channel Cav1.1 at 3.6 Å resolution. Nature 2016, 537, 191–196. [Google Scholar] [CrossRef]
- Carpenter, E.P.; Beis, K.; Cameron, A.D.; Iwata, S. Overcoming the challenges of membrane protein crystallography. Curr. Opin. Struct. Biol. 2008, 18, 581–586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, H.; Zhou, Q.; Pan, X.; Li, Z.; Wu, J.; Yan, N. Structure of a eukaryotic voltage-gated sodium channel at near-atomic resolution. Science 2017, 355, 4326. [Google Scholar] [CrossRef] [PubMed]
- Ren, D.; Navarro, B.; Xu, H.; Yue, L.; Shi, Q.; Clapham, D.E. A prokaryotic voltage-gated sodium channel. Science 2001, 294, 2372–2375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Ren, W.; DeCaen, P.; Yan, C.; Tao, X.; Tang, L.; Wang, J.; Hasegawa, K.; Kumasaka, T.; He, J.; et al. Crystal structure of an orthologue of the NaChBac voltage-gated sodium channel. Nature 2012, 486, 130–134. [Google Scholar] [CrossRef] [PubMed]
- Finol-Urdaneta, R.K.; Wang, Y.; Al-Sabi, A.; Zhao, C.; Noskov, S.Y.; French, R.J. Sodium channel selectivity and conduction: Prokaryotes have devised their own molecular strategy. J. Gen. Physiol. 2014, 143, 157–171. [Google Scholar] [CrossRef] [Green Version]
- Catterall, W.A.; Zheng, N. Deciphering voltage-gated Na(+) and Ca(2+) channels by studying prokaryotic ancestors. Trends Biochem. Sci. 2015, 40, 526–534. [Google Scholar] [CrossRef] [Green Version]
- Naylor, C.E.; Bagnéris, C.; DeCaen, P.G.; Sula, A.; Scaglione, A.; Clapham, D.E.; Wallace, B.A. Molecular basis of ion permeability in a voltage-gated sodium channel. EMBO J. 2016, 35, 820–830. [Google Scholar] [CrossRef]
- Bagneris, C.; DeCaen, P.G.; Naylor, C.E.; Pryde, D.C.; Nobeli, I.; Clapham, D.E.; Wallace, B.A. Prokaryotic NavMs channel as a structural and functional model for eukaryotic sodium channel antagonism. Proc. Natl. Acad. Sci. USA 2014, 111, 8428–8433. [Google Scholar] [CrossRef] [Green Version]
- Guardiani, C.; Rodger, P.M.; Fedorenko, O.A.; Roberts, S.K.; Khovanov, I.A. Sodium binding sites and permeation mechanism in the NaChBac channel: A molecular dynamics study. J. Chem. Theory Comput. 2017, 13, 1389–1400. [Google Scholar] [CrossRef] [Green Version]
- Yue, L.; Navarro, B.; Ren, D.; Ramos, A.; Clapham, D.E. The cation selectivity filter of the bacterial sodium channel, NaChBac. J. Gen. Physiol. 2002, 120, 845–853. [Google Scholar] [CrossRef] [Green Version]
- Tang, L.; Gamal El-Din, T.M.; Payandeh, J.; Martinez, G.Q.; Heard, T.M.; Scheuer, T.; Zheng, N.; Catterall, W.A. Structural basis for Ca2+ selectivity of a voltage-gated calcium channel. Nature 2014, 505, 56–61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guardiani, C.; Fedorenko, O.A.; Khovanov, I.A.; Roberts, S.K. Different roles for aspartates and glutamates for cation permeation in bacterial sodium channels. Biochim. Biophys. Acta (BBA)-Biomembr. 2019, 1861, 495–503. [Google Scholar] [CrossRef] [PubMed]
- Fedorenko, O.A.; Kaufman, I.K.; Gibby, W.A.T.; Barabash, M.L.; Luchinsky, D.G.; Roberts, S.K.; McClintock, P.V.E. Ionic coulomb blockade and the determinants of selectivity in the NaChBac bacterial sodium channel. Biochim. Biophys. Acta (BBA)-Biomembr. 2020, 1862, 183301. [Google Scholar] [CrossRef] [PubMed]
- Kaufman, I.K.; McClintock, P.V.E.; Eisenberg, R.S. Coulomb blockade model of permeation and selectivity in biological ion channels. New J. Phys. 2015, 17, 083021. [Google Scholar] [CrossRef]
- Kaufman, I.K.; Fedorenko, O.A.; Luchinsky, D.G.; Gibby, W.A.T.; Roberts, S.K.; McClintock, P.V.E.; Eisenberg, R.S. Ionic Coulomb blockade and anomalous mole fraction effect in the NaChBac bacterial ion channel and its charge-varied mutants. Nonlinear Biomed. Phys. 2017, 5, 4–12. [Google Scholar] [CrossRef] [Green Version]
- Payandeh, J.; Scheuer, T.; Zheng, N.; Catterall, W.A. The crystal structure of a voltage-gated sodium channel. Nature 2011, 475, 353–358. [Google Scholar] [CrossRef] [Green Version]
- Payandeh, J.; Gamal El-Din, T.M.; Scheuer, T.; Zheng, N.; Catterall, W.A. Crystal structure of a voltage-gated sodium channel in two potentially inactivated states. Nature 2012, 486, 135–139. [Google Scholar] [CrossRef] [Green Version]
- Bagnéris, C.; Decaen, P.G.; Hall, B.A.; Naylor, C.E.; Clapham, D.E.; Kay, C.W.; Wallace, B.A. Role of the C-terminal domain in the structure and function of tetrameric sodium channels. Nat. Commun. 2013, 4, 2465. [Google Scholar] [CrossRef] [Green Version]
- Shaya, D.; Findeisen, F.; Abderemane-Ali, F.; Arrigoni, C.; Wong, S.; Nurva, S.R.; Loussouarn, G.; Minor, D.L., Jr. Structure of a prokaryotic sodium channel pore reveals essential gating elements and an outer ion binding site common to eukaryotic channels. J. Mol. Biol. 2014, 426, 467–483. [Google Scholar] [CrossRef] [Green Version]
- Bagnéris, C.; Naylor, C.E.; McCusker, E.C.; Wallace, B.A. Structural model of the open-closed-inactivated cycle of prokaryotic voltage-gated sodium channels. J. Gen. Physiol. 2015, 145, 5–16. [Google Scholar] [CrossRef] [Green Version]
- McCusker, E.C.; Bagnéris, C.; Naylor, C.E.; Cole, A.R.; D’Avanzo, N.; Nichols, C.G.; Wallace, B.A. Structure of a bacterial voltage-gated sodium channel pore reveals mechanisms of opening and closing. Nat. Commun. 2012, 3, 1102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dudev, T.; Lim, C. Why voltage-gated Ca2+ and bacterial Na+ channels with the same EEEE motif in their selectivity filters confer opposite metal selectivity. Phys. Chem. Chem. Phys. 2012, 14, 12451–12456. [Google Scholar] [CrossRef] [PubMed]
- Furini, S.; Barbini, P.; Domene, C. Effects of the protonation state of the EEEE motif of a bacterial Na+ channel on conduction and pore structure. Biophys. J. 2014, 106, 2175–2183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boiteux, C.; Flood, E.; Allen, T.W. Comparison of permeation mechanisms in sodium-selective ion channels. Neurosci. Lett. 2019, 700, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Flood, E.; Boiteux, C.; Allen, T.W. Selective ion permeation involves complexation with carboxylates and lysine in a model human sodium channel. PLoS Comput. Biol. 2018, 14, e1006398. [Google Scholar] [CrossRef] [Green Version]
- Kaufman, I.K.; Luchinsky, D.G.; Gibby, W.A.T.; McClintock, P.V.E.; Eisenberg, R.S. Putative resolution of the EEEE selectivity paradox in L-type Ca2+ and bacterial Na+ biological ion channels. J. Stat. Mech. 2016, 054027. [Google Scholar] [CrossRef] [Green Version]
- Cheng, R.C.; Tikhonov, D.B.; Zhorov, B.S. Structural modeling of calcium binding in the selectivity filter of the L-type calcium channel. Eur. Biophys. J. 2010, 39, 839853. [Google Scholar] [CrossRef]
- Sun, H.; Zheng, Z.; Fedorenko, O.A.; Roberts, S.K. Covalent linkage of bacterial voltage-gated sodium channels. BMC Biophysics. 2019, 12, 1. [Google Scholar] [CrossRef]
- Neher, E. Correction for liquid junction potentials in patch clamp experiments. Methods Enzymol. 1992, 207, 123–131. [Google Scholar] [CrossRef]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef]
- Wu, E.L.; Cheng, X.; Jo, S.; Rui, H.; Song, K.C.; Dávila-Contreras, E.M.; Qi, Y.; Lee, J.; Monje-Galvan, V.; Venable, R.M.; et al. CHARMM-GUI membrane builder toward realistic biological membrane simulations. J. Comput. Chem. 2014, 35, 1997–2004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kale, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [Green Version]
- Dickson, C.J.; Madej, B.D.; Skjevik, A.A.; Betz, R.M.; Teigen, K.; Gould, I.R.; Walker, R.C. Lipid14: The amber lipid force field. J. Chem. Theory Comput. 2014, 10, 865–879. [Google Scholar] [CrossRef] [PubMed]
- Ulmschneider, M.B.; Bagneris, C.; McCusker, E.C.; DeCaen, P.G.; Delling, M.; Clapham, D.E.; Ulmschneider, J.P.; Wallace, B.A. Molecular dynamics of ion transport through the open conformation of a bacterial voltage-gated sodium channel. Proc. Natl. Acad. Sci. USA 2013, 110, 6364–6369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Zhu, F. Collective diffusion model for ion conduction through microscopic channels. Biophys. J. 2013, 104, 368–376. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Scheuer, T.; Catterall, W.A. Reversed voltage-dependent gating of a bacterial sodium channel with proline substitutions in the S6 transmembrane segment. Proc. Natl. Acad. Sci. USA 2004, 101, 17873–17878. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Yarov-Yarovoy, V.; Scheuer, T.; Catterall, W.A. A gating hinge in sodium channels; a molecular switch for electrical signaling. Neuron 2004, 41, 859–865. [Google Scholar] [CrossRef] [Green Version]
- Guardiani, C.; Fedorenko, O.A.; Roberts, S.K.; Khovanov, I.A. On the selectivity of the NaChBac channel: An integrated computational and experimental analysis of Na+ and Ca2+ permeation. Phys. Chem. Chem. Phys. 2017, 19, 29840–29854. [Google Scholar] [CrossRef] [Green Version]
- Ke, S.; Zangerl, E.M.; Stary-Weinzinger, A. Distinct interactions of Na+ and Ca2+ ions with the selectivity filter of the bacterial sodium channel NavAb. Biochem. Biophys. Res. Commun. 2013, 430, 1272–1276. [Google Scholar] [CrossRef]




| cDNA Population Transfected into CHO Cells | LASWAS | LASWAS LESWAS (3:1) | LASWAS LESWAS (1:1) | LASWAS LESWAS (1:3) | LESWAS | LESWAS LEDWAS (3:1) | LESWAS LEDWAS (1:1) | LESWAS LEDWAS (1:3) | LEDWAS |
|---|---|---|---|---|---|---|---|---|---|
| Probabilities and Qf values for tetramer formation | Qf = 0: 100% | Qf = 0; 32% Qf = −1; 42% Qf = −2; 21% Qf = −3; 4% Qf = −4; 0.3% | Qf = 0; 6.2% Qf = −1; 25% Qf = −2; 37% Qf = −3; 25% Qf = −4; 6.2% | Qf = 0; 0.3% Qf = −1; 4% Qf = −2; 21% Qf = −3; 42% Qf = −4; 32% | Qf = −4; 100% | Qf = −4; 32% Qf = −5; 42% Qf = −6; 21% Qf = −7; 4% Qf = −8; 0.3% | Qf = −4; 6.2% Qf = −5; 25% Qf = −6; 37% Qf = −7; 25% Qf = −8; 6.2% | Qf = −4; 0.3% Qf = −5; 4% Qf = −6; 21% Qf = −7; 42% Qf = −8; 32% | Qf = −8; 100% |
| Species | Ion | Conductance (pS) | Currents (pA; −20 mV) |
|---|---|---|---|
| EEEE | Ca2+ | 1.69 | −0.033 |
| EEEED | Ca2+ | 4.87 | −0.097 |
| EEEE | Na+ | 23.06 | −0.46 |
| EEEED | Na+ | 35.37 | −0.70 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fedorenko, O.A.; Khovanov, I.A.; Roberts, S.K.; Guardiani, C. Changes in Ion Selectivity Following the Asymmetrical Addition of Charge to the Selectivity Filter of Bacterial Sodium Channels. Entropy 2020, 22, 1390. https://0-doi-org.brum.beds.ac.uk/10.3390/e22121390
Fedorenko OA, Khovanov IA, Roberts SK, Guardiani C. Changes in Ion Selectivity Following the Asymmetrical Addition of Charge to the Selectivity Filter of Bacterial Sodium Channels. Entropy. 2020; 22(12):1390. https://0-doi-org.brum.beds.ac.uk/10.3390/e22121390
Chicago/Turabian StyleFedorenko, Olena A., Igor A. Khovanov, Stephen K. Roberts, and Carlo Guardiani. 2020. "Changes in Ion Selectivity Following the Asymmetrical Addition of Charge to the Selectivity Filter of Bacterial Sodium Channels" Entropy 22, no. 12: 1390. https://0-doi-org.brum.beds.ac.uk/10.3390/e22121390