Fast Approximations of the Jeffreys Divergence between Univariate Gaussian Mixtures via Mixture Conversions to Exponential-Polynomial Distributions
Abstract
:1. Introduction
1.1. Statistical Mixtures and Statistical Divergences
1.2. Jeffreys Divergence between Densities of an Exponential Family
1.3. A Simple Approximation Heuristic
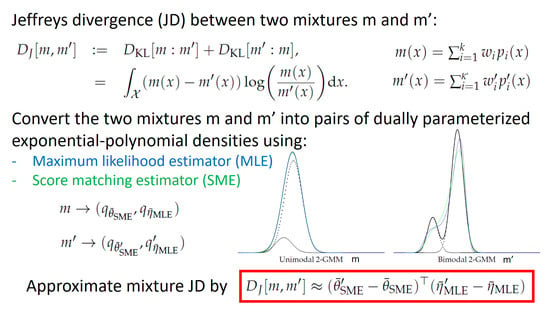
- Simplify GMMs into , and approximately convert the ’s into ’s. Then approximate the Jeffreys divergence as
- Simplify GMMs into , and approximately convert the ’s into ’s. Then approximate the Jeffreys divergence as
1.4. Contributions and Paper Outline
- We explain how to convert any continuous density (including GMMs) into a polynomial exponential density in Section 2 using integral-based extensions of the Maximum Likelihood Estimator [22] (MLE estimates in the moment parameter space H, Theorem 1 and Corollary 1) and the Score Matching Estimator [27] (SME estimates in the natural parameter space , Theorem 3). We show a connection between SME and the Moment Linear System Estimator [28] (MLSE).
- We show how to approximate the Jeffreys divergence between GMMs using a pair of natural/moment parameter PED conversion and present experimental results that display a gain of several orders of magnitude of performance when compared to the vanilla Monte Carlo estimator in Section 4. We observe that the quality of the approximations depend on the number of modes of the GMMs [43]. However, calculating or counting the modes of a GMM is a difficult problem in its own [43].
2. Converting Finite Mixtures to Exponential Family Densities
2.1. Conversion Using the Moment Parameterization (MLE)
2.2. Converting to a PEF Using the Natural Parameterization (SME)
Integral-Based Score Matching Estimator (SME)
- Generic solution: It can be shown that for exponential families [47], we obtain the following solution:whereis a symmetric matrix, andis a D-dimensional column vector.
- Solution instantiated for polynomial exponential families:For polynomial exponential families of order D, we have and , and therefore, we haveandwhere denotes the l-th raw moment of distribution (with the convention that ). For a probability density function , we have .Thus, the integral-based SME of a density r is:For example, matrix is
- Faster PEF solutions using Hankel matrices:The method of Cobb et al. [28] (1983) anticipated the Score Matching method of Hyvärinen (2005). It can be derived from Stein’s lemma for exponential families [50]. The integral-based Score Matching method is consistent, i.e., if , then : The probabilistic proof for is reported as Theorem 2 of [28]. The integral-based proof is based on the property that arbitrary order partial mixed derivatives can be obtained from higher-order partial derivatives with respect to [29]:where .The complexity of the direct SME method is as it requires the inverse of the -dimensional matrix .We show how to lower this complexity by reporting an equivalent method (originally presented in [28]) that relies on recurrence relationships between the moments of for PEDs. Recall that denotes the l-th raw moment .Let denote the symmetric matrix with (with ), and the D-dimensional vector with . We solve the system to obtain . We then obtain the natural parameter from the vector asNow, if we inspect matrix , we find that matrix is a Hankel matrix: A Hankel matrix has constant anti-diagonals and can be inverted in quadratic-time [51,52] instead of cubic time for a general matrix. (The inverse of a Hankel matrix is a Bezoutian matrix [53].) Moreover, a Hankel matrix can be stored using linear memory (store coefficients) instead of quadratic memory of regular matrices.For example, matrix is:and requires only coefficients to be stored instead of . The order-d moment matrix isis a Hankel matrix stored using coefficients:In statistics, those matrices are called moment matrices and well-studied [54,55,56]. The variance of a random variable X can be expressed as the determinant of the order-2 moment matrix:This observation yields a generalization of the notion of variance to random variables: . The variance can be expressed as for . See [57] (Chapter 5) for a detailed description related to U-statistics.For GMMs r, the raw moments to build matrix can be calculated in closed-form, as explained in Section 2.4.
2.3. Converting Numerically Moment Parameters from/to Natural Parameters
2.3.1. Converting Moment Parameters to Natural Parameters Using Maximum Entropy
2.3.2. Converting Natural Parameters to Moment Parameters
2.4. Raw Non-Central Moments of Normal Distributions and GMMs
3. Goodness-of-Fit between GMMs and PEDs: Higher Order Hyvärinen Divergences
4. Experiments: Jeffreys Divergence between Mixtures
5. Conclusions and Perspectives
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jeffreys, H. An invariant form for the prior probability in estimation problems. Proc. R. Soc. Lond. Ser. A Math. Phys. Sci. 1946, 186, 453–461. [Google Scholar]
- McLachlan, G.J.; Basford, K.E. Mixture Models: Inference and Applications to Clustering; M. Dekker: New York, NY, USA, 1988; Volume 38. [Google Scholar]
- Pearson, K. Contributions to the mathematical theory of evolution. Philos. Trans. R. Soc. Lond. A 1894, 185, 71–110. [Google Scholar]
- Seabra, J.C.; Ciompi, F.; Pujol, O.; Mauri, J.; Radeva, P.; Sanches, J. Rayleigh mixture model for plaque characterization in intravascular ultrasound. IEEE Trans. Biomed. Eng. 2011, 58, 1314–1324. [Google Scholar] [CrossRef] [PubMed]
- Kullback, S. Information Theory and Statistics; Courier Corporation: North Chelmsford, MA, USA, 1997. [Google Scholar]
- Cover, T.M. Elements of Information Theory; John Wiley & Sons: Hoboken, NJ, USA, 1999. [Google Scholar]
- Vitoratou, S.; Ntzoufras, I. Thermodynamic Bayesian model comparison. Stat. Comput. 2017, 27, 1165–1180. [Google Scholar] [CrossRef] [Green Version]
- Kannappan, P.; Rathie, P. An axiomatic characterization of J-divergence. In Transactions of the Tenth Prague Conference on Information Theory, Statistical Decision Functions, Random Processes; Springer: Dordrecht, The Netherlands, 1988; pp. 29–36. [Google Scholar]
- Burbea, J. J-Divergences and related concepts. Encycl. Stat. Sci. 2004. [Google Scholar] [CrossRef]
- Tabibian, S.; Akbari, A.; Nasersharif, B. Speech enhancement using a wavelet thresholding method based on symmetric Kullback–Leibler divergence. Signal Process. 2015, 106, 184–197. [Google Scholar] [CrossRef]
- Veldhuis, R. The centroid of the symmetrical Kullback-Leibler distance. IEEE Signal Process. Lett. 2002, 9, 96–99. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, F. Jeffreys centroids: A closed-form expression for positive histograms and a guaranteed tight approximation for frequency histograms. IEEE Signal Process. Lett. 2013, 20, 657–660. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, S.; Yamazaki, K.; Aoyagi, M. Kullback information of normal mixture is not an analytic function. IEICE Tech. Rep. Neurocomput. 2004, 104, 41–46. [Google Scholar]
- Cui, S.; Datcu, M. Comparison of Kullback-Leibler divergence approximation methods between Gaussian mixture models for satellite image retrieval. In Proceedings of the 2015 IEEE International Geoscience and Remote Sensing Symposium (IGARSS), Milan, Italy, 26–31 July 2015; pp. 3719–3722. [Google Scholar]
- Cui, S. Comparison of approximation methods to Kullback–Leibler divergence between Gaussian mixture models for satellite image retrieval. Remote Sens. Lett. 2016, 7, 651–660. [Google Scholar] [CrossRef] [Green Version]
- Sreekumar, S.; Zhang, Z.; Goldfeld, Z. Non-asymptotic Performance Guarantees for Neural Estimation of f-Divergences. In Proceedings of the International Conference on Artificial Intelligence and Statistics (PMLR 2021), San Diego, CA, USA, 18–24 July 2021; pp. 3322–3330. [Google Scholar]
- Durrieu, J.L.; Thiran, J.P.; Kelly, F. Lower and upper bounds for approximation of the Kullback-Leibler divergence between Gaussian mixture models. In Proceedings of the 2012 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Kyoto, Japan, 25–30 March 2012; pp. 4833–4836. [Google Scholar]
- Nielsen, F.; Sun, K. Guaranteed bounds on information-theoretic measures of univariate mixtures using piecewise log-sum-exp inequalities. Entropy 2016, 18, 442. [Google Scholar] [CrossRef] [Green Version]
- Jenssen, R.; Principe, J.C.; Erdogmus, D.; Eltoft, T. The Cauchy–Schwarz divergence and Parzen windowing: Connections to graph theory and Mercer kernels. J. Frankl. Inst. 2006, 343, 614–629. [Google Scholar] [CrossRef]
- Liu, M.; Vemuri, B.C.; Amari, S.i.; Nielsen, F. Shape retrieval using hierarchical total Bregman soft clustering. IEEE Trans. Pattern Anal. Mach. Intell. 2012, 34, 2407–2419. [Google Scholar]
- Robert, C.; Casella, G. Monte Carlo Statistical Methods; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2013. [Google Scholar]
- Barndorff-Nielsen, O. Information and Exponential Families: In Statistical Theory; John Wiley & Sons: Hoboken, NJ, USA, 2014. [Google Scholar]
- Azoury, K.S.; Warmuth, M.K. Relative loss bounds for on-line density estimation with the exponential family of distributions. Mach. Learn. 2001, 43, 211–246. [Google Scholar] [CrossRef] [Green Version]
- Banerjee, A.; Merugu, S.; Dhillon, I.S.; Ghosh, J. Clustering with Bregman divergences. J. Mach. Learn. Res. 2005, 6, 1705–1749. [Google Scholar]
- Bregman, L.M. The relaxation method of finding the common point of convex sets and its application to the solution of problems in convex programming. USSR Comput. Math. Math. Phys. 1967, 7, 200–217. [Google Scholar] [CrossRef]
- Nielsen, F.; Nock, R. Sided and symmetrized Bregman centroids. IEEE Trans. Inf. Theory 2009, 55, 2882–2904. [Google Scholar] [CrossRef] [Green Version]
- Hyvärinen, A. Estimation of non-normalized statistical models by score matching. J. Mach. Learn. Res. 2005, 6, 695–709. [Google Scholar]
- Cobb, L.; Koppstein, P.; Chen, N.H. Estimation and moment recursion relations for multimodal distributions of the exponential family. J. Am. Stat. Assoc. 1983, 78, 124–130. [Google Scholar] [CrossRef]
- Hayakawa, J.; Takemura, A. Estimation of exponential-polynomial distribution by holonomic gradient descent. Commun. Stat.-Theory Methods 2016, 45, 6860–6882. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, F.; Nock, R. MaxEnt upper bounds for the differential entropy of univariate continuous distributions. IEEE Signal Process. Lett. 2017, 24, 402–406. [Google Scholar] [CrossRef]
- Matz, A.W. Maximum likelihood parameter estimation for the quartic exponential distribution. Technometrics 1978, 20, 475–484. [Google Scholar] [CrossRef]
- Barron, A.R.; Sheu, C.H. Approximation of density functions by sequences of exponential families. Ann. Stat. 1991, 19, 1347–1369, Correction in 1991, 19, 2284–2284. [Google Scholar]
- O’toole, A. A method of determining the constants in the bimodal fourth degree exponential function. Ann. Math. Stat. 1933, 4, 79–93. [Google Scholar] [CrossRef]
- Aroian, L.A. The fourth degree exponential distribution function. Ann. Math. Stat. 1948, 19, 589–592. [Google Scholar] [CrossRef]
- Zellner, A.; Highfield, R.A. Calculation of maximum entropy distributions and approximation of marginal posterior distributions. J. Econom. 1988, 37, 195–209. [Google Scholar] [CrossRef]
- McCullagh, P. Exponential mixtures and quadratic exponential families. Biometrika 1994, 81, 721–729. [Google Scholar] [CrossRef]
- Mead, L.R.; Papanicolaou, N. Maximum entropy in the problem of moments. J. Math. Phys. 1984, 25, 2404–2417. [Google Scholar] [CrossRef] [Green Version]
- Armstrong, J.; Brigo, D. Stochastic filtering via L2 projection on mixture manifolds with computer algorithms and numerical examples. arXiv 2013, arXiv:1303.6236. [Google Scholar]
- Efron, B.; Hastie, T. Computer Age Statistical Inference; Cambridge University Press: Cambridge, UK, 2016; Volume 5. [Google Scholar]
- Pinsker, M. Information and Information Stability of Random Variables and Processes (Translated and Annotated by Amiel Feinstein); Holden-Day Inc.: San Francisco, CA, USA, 1964. [Google Scholar]
- Fedotov, A.A.; Harremoës, P.; Topsoe, F. Refinements of Pinsker’s inequality. IEEE Trans. Inf. Theory 2003, 49, 1491–1498. [Google Scholar] [CrossRef]
- Amari, S. Information Geometry and Its Applications; Applied Mathematical Sciences; Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar]
- Carreira-Perpinan, M.A. Mode-finding for mixtures of Gaussian distributions. IEEE Trans. Pattern Anal. Mach. Intell. 2000, 22, 1318–1323. [Google Scholar] [CrossRef] [Green Version]
- Brown, L.D. Fundamentals of statistical exponential families with applications in statistical decision theory. Lect. Notes-Monogr. Ser. 1986, 9, 1–279. [Google Scholar]
- Pelletier, B. Informative barycentres in statistics. Ann. Inst. Stat. Math. 2005, 57, 767–780. [Google Scholar] [CrossRef]
- Améndola, C.; Drton, M.; Sturmfels, B. Maximum likelihood estimates for Gaussian mixtures are transcendental. In Proceedings of the International Conference on Mathematical Aspects of Computer and Information Sciences, Berlin, Germany, 11–13 November 2015; pp. 579–590. [Google Scholar]
- Hyvärinen, A. Some extensions of score matching. Comput. Stat. Data Anal. 2007, 51, 2499–2512. [Google Scholar] [CrossRef] [Green Version]
- Otto, F.; Villani, C. Generalization of an inequality by Talagrand and links with the logarithmic Sobolev inequality. J. Funct. Anal. 2000, 173, 361–400. [Google Scholar] [CrossRef] [Green Version]
- Toscani, G. Entropy production and the rate of convergence to equilibrium for the Fokker-Planck equation. Q. Appl. Math. 1999, 57, 521–541. [Google Scholar] [CrossRef] [Green Version]
- Hudson, H.M. A natural identity for exponential families with applications in multiparameter estimation. Ann. Stat. 1978, 6, 473–484. [Google Scholar] [CrossRef]
- Trench, W.F. An algorithm for the inversion of finite Hankel matrices. J. Soc. Ind. Appl. Math. 1965, 13, 1102–1107. [Google Scholar] [CrossRef]
- Heinig, G.; Rost, K. Fast algorithms for Toeplitz and Hankel matrices. Linear Algebra Its Appl. 2011, 435, 1–59. [Google Scholar] [CrossRef] [Green Version]
- Fuhrmann, P.A. Remarks on the inversion of Hankel matrices. Linear Algebra Its Appl. 1986, 81, 89–104. [Google Scholar] [CrossRef] [Green Version]
- Lindsay, B.G. On the determinants of moment matrices. Ann. Stat. 1989, 17, 711–721. [Google Scholar] [CrossRef]
- Lindsay, B.G. Moment matrices: Applications in mixtures. Ann. Stat. 1989, 17, 722–740. [Google Scholar] [CrossRef]
- Provost, S.B.; Ha, H.T. On the inversion of certain moment matrices. Linear Algebra Its Appl. 2009, 430, 2650–2658. [Google Scholar] [CrossRef]
- Serfling, R.J. Approximation Theorems of Mathematical Statistics; John Wiley & Sons: Hoboken, NJ, USA, 2009; Volume 162. [Google Scholar]
- Mohammad-Djafari, A. A. A Matlab program to calculate the maximum entropy distributions. In Maximum Entropy and Bayesian Methods; Springer: Berlin/Heidelberg, Germany, 1992; pp. 221–233. [Google Scholar]
- Karlin, S. Total Positivity; Stanford University Press: Redwood City, CA, USA, 1968; Volume 1. [Google Scholar]
- von Neumann, J. Various Techniques Used in Connection with Random Digits. In Monte Carlo Method; National Bureau of Standards Applied Mathematics Series; Householder, A.S., Forsythe, G.E., Germond, H.H., Eds.; US Government Printing Office: Washington, DC, USA, 1951; Volume 12, Chapter 13; pp. 36–38. [Google Scholar]
- Flury, B.D. Acceptance-rejection sampling made easy. SIAM Rev. 1990, 32, 474–476. [Google Scholar] [CrossRef]
- Rohde, D.; Corcoran, J. MCMC methods for univariate exponential family models with intractable normalization constants. In Proceedings of the 2014 IEEE Workshop on Statistical Signal Processing (SSP), Gold Coast, Australia, 29 June–2 July 2014; pp. 356–359. [Google Scholar]
- Barr, D.R.; Sherrill, E.T. Mean and variance of truncated normal distributions. Am. Stat. 1999, 53, 357–361. [Google Scholar]
- Amendola, C.; Faugere, J.C.; Sturmfels, B. Moment Varieties of Gaussian Mixtures. J. Algebr. Stat. 2016, 7, 14–28. [Google Scholar] [CrossRef] [Green Version]
- Fujisawa, H.; Eguchi, S. Robust parameter estimation with a small bias against heavy contamination. J. Multivar. Anal. 2008, 99, 2053–2081. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, F.; Nock, R. Patch matching with polynomial exponential families and projective divergences. In Proceedings of the International Conference on Similarity Search and Applications, Tokyo, Japan, 24–26 October 2016; pp. 109–116. [Google Scholar]
- Yang, Y.; Martin, R.; Bondell, H. Variational approximations using Fisher divergence. arXiv 2019, arXiv:1905.05284. [Google Scholar]
- Kostrikov, I.; Fergus, R.; Tompson, J.; Nachum, O. Offline reinforcement learning with Fisher divergence critic regularization. In Proceedings of the International Conference on Machine Learning (PMLR 2021), online, 7–8 June 2021; pp. 5774–5783. [Google Scholar]
- Elkhalil, K.; Hasan, A.; Ding, J.; Farsiu, S.; Tarokh, V. Fisher Auto-Encoders. In Proceedings of the International Conference on Artificial Intelligence and Statistics (PMLR 2021), San Diego, CA, USA, 13–15 April 2021; pp. 352–360. [Google Scholar]
- Améndola, C.; Engström, A.; Haase, C. Maximum number of modes of Gaussian mixtures. Inf. Inference J. IMA 2020, 9, 587–600. [Google Scholar] [CrossRef]
- Aprausheva, N.; Mollaverdi, N.; Sorokin, S. Bounds for the number of modes of the simplest Gaussian mixture. Pattern Recognit. Image Anal. 2006, 16, 677–681. [Google Scholar] [CrossRef] [Green Version]
- Aprausheva, N.; Sorokin, S. Exact equation of the boundary of unimodal and bimodal domains of a two-component Gaussian mixture. Pattern Recognit. Image Anal. 2013, 23, 341–347. [Google Scholar] [CrossRef]
- Xiao, Y.; Shah, M.; Francis, S.; Arnold, D.L.; Arbel, T.; Collins, D.L. Optimal Gaussian mixture models of tissue intensities in brain MRI of patients with multiple-sclerosis. In Proceedings of the International Workshop on Machine Learning in Medical Imaging, Beijing, China, 20 September 2010; pp. 165–173. [Google Scholar]
- Bilik, I.; Khomchuk, P. Minimum divergence approaches for robust classification of ground moving targets. IEEE Trans. Aerosp. Electron. Syst. 2012, 48, 581–603. [Google Scholar] [CrossRef]
- Alippi, C.; Boracchi, G.; Carrera, D.; Roveri, M. Change Detection in Multivariate Datastreams: Likelihood and Detectability Loss. In Proceedings of the Twenty-Fifth International Joint Conference on Artificial Intelligence, IJCAI 2016, New York, NY, USA, 9–15 July 2016. [Google Scholar]
- Eguchi, S.; Komori, O.; Kato, S. Projective power entropy and maximum Tsallis entropy distributions. Entropy 2011, 13, 1746–1764. [Google Scholar] [CrossRef]
- Orjebin, E. A Recursive Formula for the Moments of a Truncated Univariate Normal Distribution. 2014, Unpublished note.
- Del Castillo, J. The singly truncated normal distribution: A non-steep exponential family. Ann. Inst. Stat. Math. 1994, 46, 57–66. [Google Scholar] [CrossRef] [Green Version]









| k | D | Average Error | Maximum Error | Speed-Up |
|---|---|---|---|---|
| 2 | 4 | 0.1180799978221536 | 0.9491425404132259 | 2008.2323536011806 |
| 3 | 6 | 0.12533811294546526 | 1.9420608151988419 | 1010.4917042114389 |
| 4 | 8 | 0.10198448868508087 | 5.290871019594698 | 474.5135294829539 |
| 5 | 10 | 0.06336388579897352 | 3.8096955246161848 | 246.38780782640987 |
| 6 | 12 | 0.07145257192133717 | 1.0125283726458822 | 141.39097909641052 |
| 7 | 14 | 0.10538875853178625 | 0.8661463142793943 | 88.62985036546912 |
| 8 | 16 | 0.4150905507007969 | 0.4150905507007969 | 58.72277575395611 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nielsen, F. Fast Approximations of the Jeffreys Divergence between Univariate Gaussian Mixtures via Mixture Conversions to Exponential-Polynomial Distributions. Entropy 2021, 23, 1417. https://0-doi-org.brum.beds.ac.uk/10.3390/e23111417
Nielsen F. Fast Approximations of the Jeffreys Divergence between Univariate Gaussian Mixtures via Mixture Conversions to Exponential-Polynomial Distributions. Entropy. 2021; 23(11):1417. https://0-doi-org.brum.beds.ac.uk/10.3390/e23111417
Chicago/Turabian StyleNielsen, Frank. 2021. "Fast Approximations of the Jeffreys Divergence between Univariate Gaussian Mixtures via Mixture Conversions to Exponential-Polynomial Distributions" Entropy 23, no. 11: 1417. https://0-doi-org.brum.beds.ac.uk/10.3390/e23111417