HIV Resistance Prediction to Reverse Transcriptase Inhibitors: Focus on Open Data
Abstract
:1. Introduction
2. Results and Discussion
2.1. Experimental Methods of HIV-1 Resistance Detection and Concordance between Their Results
2.2. HIV Sequences Repositories
2.2.1. NCBI Nucleotide (GenBank) and NCBI Protein
2.2.2. Los Alamos HIV Sequence Database
2.2.3. The HIV Oligonucleotide Database (HIVoligoDB)
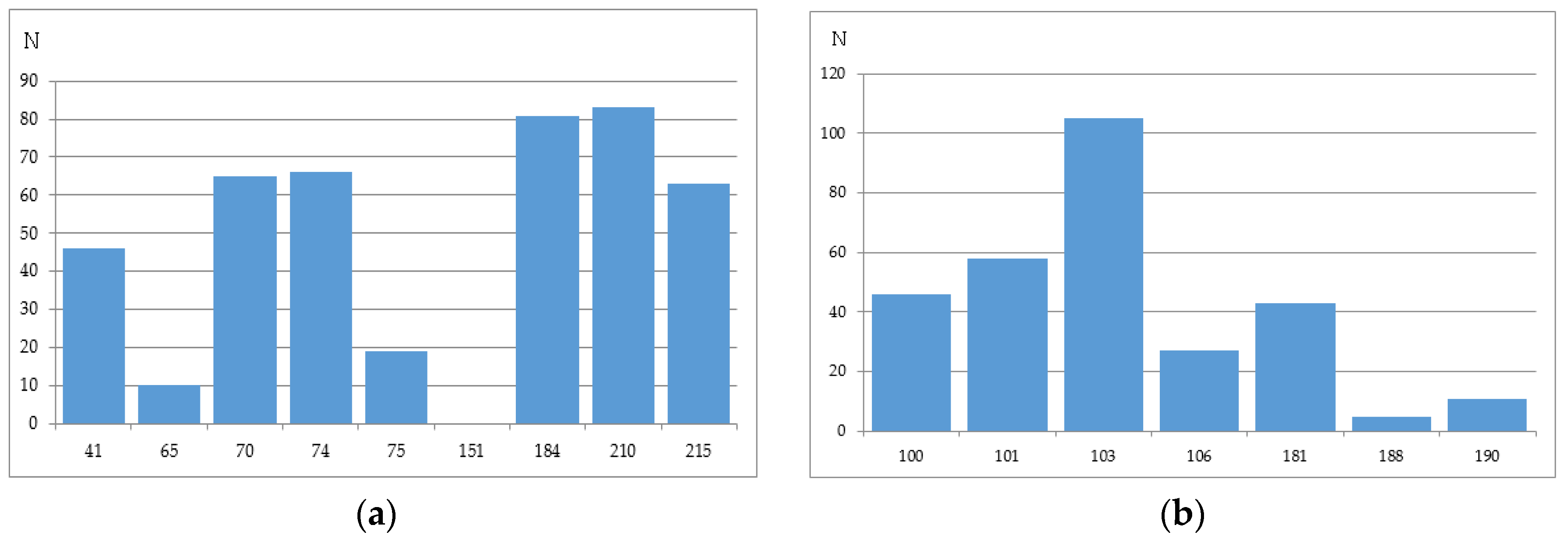
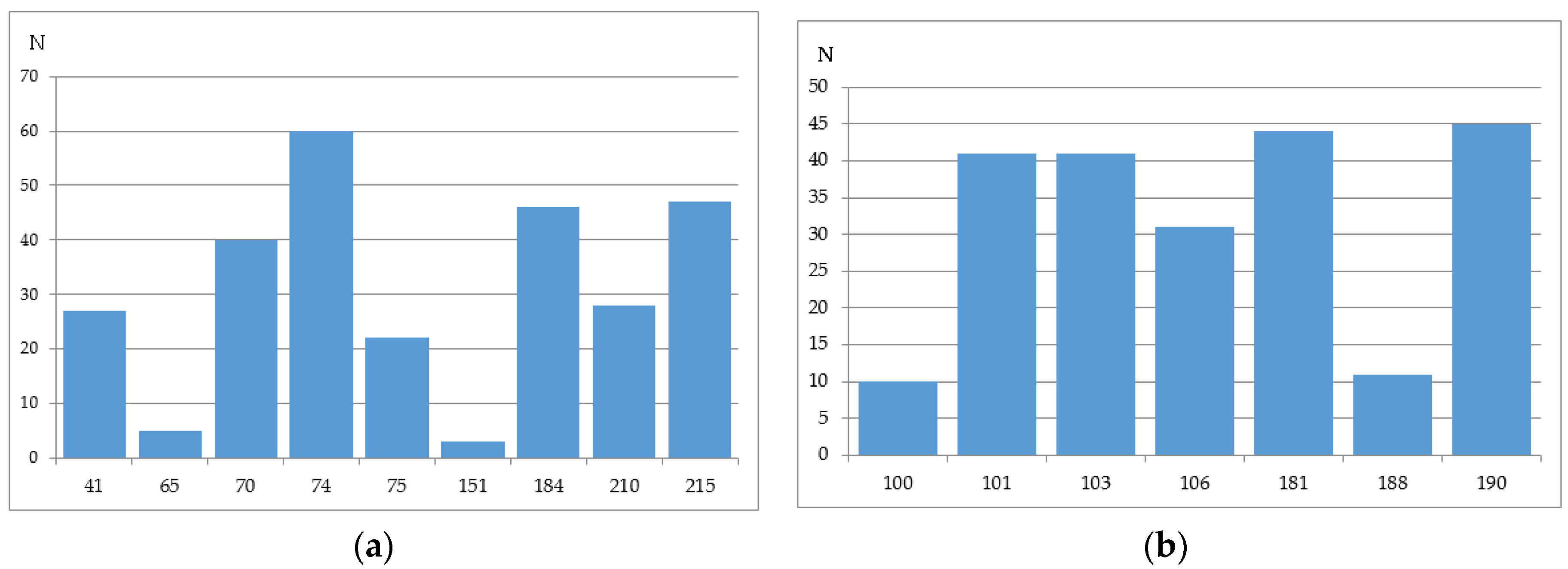
2.2.4. Stanford University HIV Drug Resistance Database
2.3. Computational Methods of HIV-1 RT Associated Resistance and the Level of Concordance between Them
2.4. Perspectives of the HIV Variants’ Open Data Use for HIV Resistance Prediction in Clinical Practice
2.5. Perspectives of the HIV Variants’ Open Data Use for New Drug Development
3. Conclusions
Supplementary Materials
Acknowledgments
Conflicts of Interest
References
- Distinto, S.; Maccioni, E.; Meleddu, R.; Corona, A.; Alcaro, S.; Tramontano, E. Molecular aspects of the RT/drug interactions. Perspective of dual inhibitors. Curr. Pharm. Des. 2013, 19, 1850–1859. [Google Scholar] [CrossRef] [PubMed]
- Hao, G.-F.; Yang, S.-G.; Yang, G.-F. Structure-based design of conformationally flexible reverse transcriptase inhibitors to combat resistant HIV. Curr. Pharm. Des. 2014, 20, 725–739. [Google Scholar] [CrossRef] [PubMed]
- Geronikaki, A.; Eleftheriou, P.; Poroikov, V. Anti-HIV Agents: Current Status and Recent Trends; Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar]
- Sinokrot, H.; Smerat, T.; Najjar, A.; Karaman, R. Advanced Prodrug Strategies in Nucleoside and Non-Nucleoside Antiviral Agents: A Review of the Recent Five Years. Molecules 2017, 22, 1736. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Li, X.; De Clercq, E.; Zhan, P.; Liu, X. Discovery of potent HIV-1 non-nucleoside reverse transcriptase inhibitors from arylthioacetanilide structural motif. Eur. J. Med. Chem. 2015, 102, 167–179. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.; Flores, J.; Kirby, K.; Neogi, U.; Sonnerborg, A.; Hachiya, A.; Das, K.; Arnold, E.; McArthur, C.; Parniak, M.; et al. Drug Resistance in Non-B Subtype HIV-1: Impact of HIV-1 Reverse Transcriptase Inhibitors. Viruses 2014, 6, 3535–3562. [Google Scholar] [CrossRef] [Green Version]
- Novitsky, V.; Wester, C.W.; DeGruttola, V.; Bussmann, H.; Gaseitsiwe, S.; Thomas, A.; Moyo, S.; Musonda, R.; Van Widenfelt, E.; Marlink, R.G.; et al. The reverse transcriptase 67N 70R 215Y genotype is the predominant TAM pathway associated with virologic failure among HIV type 1C-infected adults treated with ZDV/ddI-containing HAART in southern Africa. AIDS Res. Hum. Retrovir. 2007, 23, 868–878. [Google Scholar] [CrossRef] [PubMed]
- Mata-Marín, J.A.; Huerta-García, G.; Domínguez-Hermosillo, J.C.; Chavez-García, M.; Banda-Lara, M.I.; Nuñez-Rodríguez, N.; Cruz-Herrera, J.E.; Sandoval-Ramírez, J.L.; Martínez-Abarca, I.; Villagómez-Ruíz, A.F.; et al. Effectiveness and risk factors for virological outcome of darunavir-based therapy for treatment-experienced HIV-infected patients. AIDS Res. Ther. 2015, 12, 31. [Google Scholar] [CrossRef] [PubMed]
- Owen, A.; Rannard, S. Strengths, weaknesses, opportunities and challenges for long acting injectable therapies: Insights for applications in HIV therapy. Adv. Drug Deliv. Rev. 2016, 103, 144–156. [Google Scholar] [CrossRef]
- Iyidogan, P.; Anderson, K.S. Current perspectives on HIV-1 antiretroviral drug resistance. Viruses 2014, 6, 4095–4139. [Google Scholar] [CrossRef] [PubMed]
- Prosperi, M.C.F.; De Luca, A. Computational models for prediction of response to antiretroviral therapies. AIDS Rev. 2012, 14, 145–153. [Google Scholar] [PubMed]
- Zazzi, M.; Cozzi-Lepri, A.; Prosperi, M.C.F. Computer-Aided Optimization of Combined Anti-Retroviral Therapy for HIV: New Drugs, New Drug Targets and Drug Resistance. Curr. HIV Res. 2016, 14, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.F.; Shafer, R.W. Web resources for HIV type 1 genotypic-resistance test interpretation. Clin. Infect. Dis. 2006, 42, 1608–1618. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Picado, J.; Martínez, M.A. HIV-1 reverse transcriptase inhibitor resistance mutations and fitness: A view from the clinic and ex vivo. Virus Res. 2008, 134, 104–123. [Google Scholar] [CrossRef] [PubMed]
- Pauwels, R. Aspects of successful drug discovery and development. Antivir. Res. 2006, 71, 77–89. [Google Scholar] [CrossRef] [PubMed]
- Demeter, L.M.; D’Aquila, R.; Weislow, O.; Lorenzo, E.; Erice, A.; Fitzgibbon, J.; Shafer, R.; Richman, D.; Howard, T.M.; Zhao, Y.; et al. Interlaboratory concordance of DNA sequence analysis to detect reverse transcriptase mutations in HIV-1 proviral DNA. ACTG Sequencing Working Group. AIDS Clinical Trials Group. J. Virol. Methods 1998, 75, 93–104. [Google Scholar] [CrossRef]
- Durant, J.; Clevenbergh, P.; Halfon, P.; Delgiudice, P.; Porsin, S.; Simonet, P.; Montagne, N.; Boucher, C.A.; Schapiro, J.M.; Dellamonica, P. Drug-resistance genotyping in HIV-1 therapy: The VIRADAPT randomised controlled trial. Lancet 1999, 353, 2195–2199. [Google Scholar] [CrossRef]
- Hertogs, K.; de Béthune, M.P.; Miller, V.; Ivens, T.; Schel, P.; Van Cauwenberge, A.; Van Den Eynde, C.; Van Gerwen, V.; Azijn, H.; Van Houtte, M.; et al. A rapid method for simultaneous detection of phenotypic resistance to inhibitors of protease and reverse transcriptase in recombinant human immunodeficiency virus type 1 isolates from patients treated with antiretroviral drugs. Antimicrob. Agents Chemother. 1998, 42, 269–276. [Google Scholar] [PubMed]
- Petropoulos, C.J.; Parkin, N.T.; Limoli, K.L.; Lie, Y.S.; Wrin, T.; Huang, W.; Tian, H.; Smith, D.; Winslow, G.A.; Capon, D.J.; et al. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 2000, 44, 920–928. [Google Scholar] [CrossRef] [PubMed]
- Thomas, P.; Smart, T.G. HEK293 cell line: A vehicle for the expression of recombinant proteins. J. Pharmacol. Toxicol. Methods 2005, 51, 187–200. [Google Scholar] [CrossRef] [PubMed]
- Qari, S.H.; Respess, R.; Weinstock, H.; Beltrami, E.M.; Hertogs, K.; Larder, B.A.; Petropoulos, C.J.; Hellmann, N.; Heneine, W. Comparative analysis of two commercial phenotypic assays for drug susceptibility testing of human immunodeficiency virus type 1. J. Clin. Microbiol. 2002, 40, 31–35. [Google Scholar] [CrossRef] [PubMed]
- Torti, C.; Quiros-Roldan, E.; Keulen, W.; Scudeller, L.; Lo Caputo, S.; Boucher, C.; Castelli, F.; Mazzotta, F.; Pierotti, P.; Been-Tiktak, A.M.; et al. GenPherex Study Group of the MaSTeR Cohort Comparison between rules-based human immunodeficiency virus type 1 genotype interpretations and real or virtual phenotype: Concordance analysis and correlation with clinical outcome in heavily treated patients. J. Infect. Dis. 2003, 188, 194–201. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Samudrala, R.; Mittler, J.E. Antivirogram or phenosense: A comparison of their reproducibility and an analysis of their correlation. Antivir. Ther. 2004, 9, 703–712. [Google Scholar]
- Zhang, J.; Rhee, S.-Y.; Taylor, J.; Shafer, R.W. Comparison of the precision and sensitivity of the Antivirogram and PhenoSense HIV drug susceptibility assays. J. Acquir. Immune Defic. Syndr. 2005, 38, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Ross, L.; Boulmé, R.; Fisher, R.; Hernandez, J.; Florance, A.; Schmit, J.-C.; Williams, V. A direct comparison of drug susceptibility to HIV type 1 from antiretroviral experienced subjects as assessed by the antivirogram and PhenoSense assays and by seven resistance algorithms. AIDS Res. Hum. Retrovir. 2005, 21, 933–939. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Samudrala, R.; Mittler, J. Weak agreement between Antivirogram and Phenosense assays in predicting reduced susceptibility to antiretroviral drugs. J. Clin. Microbiol. 2004, 42, 2353–2354. [Google Scholar] [CrossRef] [PubMed]
- Shafer, R.W.; Hertogs, K.; Zolopa, A.R.; Warford, A.; Bloor, S.; Betts, B.J.; Merigan, T.C.; Harrigan, R.; Larder, B.A. High degree of interlaboratory reproducibility of human immunodeficiency virus type 1 protease and reverse transcriptase sequencing of plasma samples from heavily treated patients. J. Clin. Microbiol. 2001, 39, 1522–1529. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.W.; Shafer, R.W. HIV-1 antiretroviral resistance: Scientific principles and clinical applications. Drugs 2012, 72, e1-25. [Google Scholar] [CrossRef] [PubMed]
- Schuurman, R.; Demeter, L.; Reichelderfer, P.; Tijnagel, J.; de Groot, T.; Boucher, C. Worldwide evaluation of DNA sequencing approaches for identification of drug resistance mutations in the human immunodeficiency virus type 1 reverse transcriptase. J. Clin. Microbiol. 1999, 37, 2291–2296. [Google Scholar] [PubMed]
- Demeter, L.; Haubrich, R. International perspectives on antiretroviral resistance. Phenotypic and genotypic resistance assays: Methodology, reliability, and interpretations. J. Acquir. Immune Defic. Syndr. 2001, 26 (Suppl. 1), S3–S9. [Google Scholar] [CrossRef] [PubMed]
- Parkin, N.; Chappey, C.; Maroldo, L.; Bates, M.; Hellmann, N.S.; Petropoulos, C.J. Phenotypic and genotypic HIV-1 drug resistance assays provide complementary information. J. Acquir. Immune Defic. Syndr. 2002, 31, 128–136. [Google Scholar] [CrossRef] [PubMed]
- Benson, D.A.; Cavanaugh, M.; Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2013, 41, D36–D42. [Google Scholar] [CrossRef] [PubMed]
- Tatusova, T. Update on Genomic Databases and Resources at the National Center for Biotechnology Information. In Data Mining Techniques for the Life Sciences; Carugo, O., Eisenhaber, F., Eds.; Springer: New York, NY, USA, 2016; Volume 1415, pp. 3–30. ISBN 978-1-4939-3570-3. [Google Scholar]
- Kim, S.; Thiessen, P.A.; Bolton, E.E.; Bryant, S.H. PUG-SOAP and PUG-REST: Web services for programmatic access to chemical information in PubChem. Nucleic Acids Res. 2015, 43, W605–W611. [Google Scholar] [CrossRef] [PubMed]
- Katayama, T.; Nakao, M.; Takagi, T. TogoWS: Integrated SOAP and REST APIs for interoperable bioinformatics Web services. Nucleic Acids Res. 2010, 38, W706–W711. [Google Scholar] [CrossRef] [PubMed]
- Kuiken, C.; Korber, B.; Shafer, R.W. HIV sequence databases. AIDS Rev. 2003, 5, 52–61. [Google Scholar] [PubMed]
- Clark, S.A.; Shulman, N.S.; Bosch, R.J.; Mellors, J.W. Reverse transcriptase mutations 118I, 208Y, and 215Y cause HIV-1 hypersusceptibility to non-nucleoside reverse transcriptase inhibitors. AIDS 2006, 20, 981–984. [Google Scholar] [CrossRef] [PubMed]
- Carneiro, J.; Resende, A.; Pereira, F. The HIV oligonucleotide database (HIVoligoDB). Database 2017, 2017. [Google Scholar] [CrossRef] [PubMed]
- Rhee, S.-Y.; Gonzales, M.J.; Kantor, R.; Betts, B.J.; Ravela, J.; Shafer, R.W. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 2003, 31, 298–303. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Chopra, R.; Verdine, G.L.; Harrison, S.C. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: Implications for drug resistance. Science 1998, 282, 1669–1675. [Google Scholar] [CrossRef] [PubMed]
- Meyer, P.R.; Matsuura, S.E.; Mian, A.M.; So, A.G.; Scott, W.A. A mechanism of AZT resistance: An increase in nucleotide-dependent primer unblocking by mutant HIV-1 reverse transcriptase. Mol. Cell 1999, 4, 35–43. [Google Scholar] [CrossRef]
- Meyer, P.R.; Matsuura, S.E.; So, A.G.; Scott, W.A. Unblocking of chain-terminated primer by HIV-1 reverse transcriptase through a nucleotide-dependent mechanism. Proc. Natl. Acad. Sci. USA 1998, 95, 13471–13476. [Google Scholar] [CrossRef] [PubMed]
- Sarafianos, S.G.; Das, K.; Clark, A.D.; Ding, J.; Boyer, P.L.; Hughes, S.H.; Arnold, E. Lamivudine (3TC) resistance in HIV-1 reverse transcriptase involves steric hindrance with beta-branched amino acids. Proc. Natl. Acad. Sci. USA 1999, 96, 10027–10032. [Google Scholar] [CrossRef] [PubMed]
- Sarafianos, S.G.; Das, K.; Ding, J.; Boyer, P.L.; Hughes, S.H.; Arnold, E. Touching the heart of HIV-1 drug resistance: The fingers close down on the dNTP at the polymerase active site. Chem. Biol. 1999, 6, R137–R146. [Google Scholar] [CrossRef]
- Arion, D.; Sluis-Cremer, N.; Parniak, M.A. Mechanism by which phosphonoformic acid resistance mutations restore 3′-azido-3′-deoxythymidine (AZT) sensitivity to AZT-resistant HIV-1 reverse transcriptase. J. Biol. Chem. 2000, 275, 9251–9255. [Google Scholar] [CrossRef] [PubMed]
- Hsiou, Y.; Ding, J.; Das, K.; Clark, A.D.; Boyer, P.L.; Lewi, P.; Janssen, P.A.; Kleim, J.P.; Rösner, M.; Hughes, S.H.; Arnold, E. The Lys103Asn mutation of HIV-1 RT: A novel mechanism of drug resistance. J. Mol. Biol. 2001, 309, 437–445. [Google Scholar] [CrossRef]
- Deval, J.; Courcambeck, J.; Selmi, B.; Boretto, J.; Canard, B. Structural determinants and molecular mechanisms for the resistance of HIV-1 RT to nucleoside analogues. Curr. Drug Metab. 2004, 5, 305–316. [Google Scholar] [CrossRef] [PubMed]
- Deval, J.; White, K.L.; Miller, M.D.; Parkin, N.T.; Courcambeck, J.; Halfon, P.; Selmi, B.; Boretto, J.; Canard, B. Mechanistic basis for reduced viral and enzymatic fitness of HIV-1 reverse transcriptase containing both K65R and M184V mutations. J. Biol. Chem. 2004, 279, 509–516. [Google Scholar] [CrossRef]
- Tuske, S.; Sarafianos, S.G.; Clark, A.D.; Ding, J.; Naeger, L.K.; White, K.L.; Miller, M.D.; Gibbs, C.S.; Boyer, P.L.; Clark, P.; et al. Structures of HIV-1 RT-DNA complexes before and after incorporation of the anti-AIDS drug tenofovir. Nat. Struct. Mol. Biol. 2004, 11, 469–474. [Google Scholar] [CrossRef] [PubMed]
- Miranda, L.R.; Götte, M.; Liang, F.; Kuritzkes, D.R. The L74V mutation in human immunodeficiency virus type 1 reverse transcriptase counteracts enhanced excision of zidovudine monophosphate associated with thymidine analog resistance mutations. Antimicrob. Agents Chemother. 2005, 49, 2648–2656. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Puchhammer-Stöckl, E.; Steininger, C.; Geringer, E.; Heinz, F.X. Comparison of virtual phenotype and HIV-SEQ program (Stanford) interpretation for predicting drug resistance of HIV strains. HIV Med. 2002, 3, 200–206. [Google Scholar] [CrossRef] [PubMed]
- Brun-Vézinet, F.; Costagliola, D.; Khaled, M.A.; Calvez, V.; Clavel, F.; Clotet, B.; Haubrich, R.; Kempf, D.; King, M.; Kuritzkes, D.; et al. Clinically validated genotype analysis: Guiding principles and statistical concerns. Antivir. Ther. 2004, 9, 465–478. [Google Scholar] [PubMed]
- Van Laethem, K.; De Luca, A.; Antinori, A.; Cingolani, A.; Perna, C.F.; Vandamme, A.-M. A genotypic drug resistance interpretation algorithm that significantly predicts therapy response in HIV-1-infected patients. Antivir. Ther. 2002, 7, 123–129. [Google Scholar] [PubMed]
- Beerenwinkel, N.; Däumer, M.; Oette, M.; Korn, K.; Hoffmann, D.; Kaiser, R.; Lengauer, T.; Selbig, J.; Walter, H. Geno2pheno: Estimating phenotypic drug resistance from HIV-1 genotypes. Nucleic Acids Res. 2003, 31, 3850–3855. [Google Scholar] [CrossRef] [PubMed]
- Beerenwinkel, N.; Schmidt, B.; Walter, H.; Kaiser, R.; Lengauer, T.; Hoffmann, D.; Korn, K.; Selbig, J. Diversity and complexity of HIV-1 drug resistance: A bioinformatics approach to predicting phenotype from genotype. Proc. Natl. Acad. Sci. USA 2002, 99, 8271–8276. [Google Scholar] [CrossRef] [PubMed]
- Revell, A.D.; Wang, D.; Wood, R.; Morrow, C.; Tempelman, H.; Hamers, R.; Alvarez-Uria, G.; Streinu-Cercel, A.; Ene, L.; Wensing, A.; et al. An update to the HIV-TRePS system: The development of new computational models that do not require a genotype to predict HIV treatment outcomes. J. Antimicrob. Chemother. 2014, 69, 1104–1110. [Google Scholar] [CrossRef] [PubMed]
- Zazzi, M.; Incardona, F.; Rosen-Zvi, M.; Prosperi, M.; Lengauer, T.; Altmann, A.; Sonnerborg, A.; Lavee, T.; Schülter, E.; Kaiser, R. Predicting response to antiretroviral treatment by machine learning: The EuResist project. Intervirology 2012, 55, 123–127. [Google Scholar] [CrossRef] [PubMed]
- De Luca, A. The impact of resistance on viral fitness and its clinical implications. In Antiretroviral Resistance in Clinical Practice; Geretti, A.M., Ed.; Mediscript: London, UK, 2006; ISBN 978-0-9551669-0-7. [Google Scholar]
- Rhee, S.-Y.; Taylor, J.; Wadhera, G.; Ben-Hur, A.; Brutlag, D.L.; Shafer, R.W. Genotypic predictors of human immunodeficiency virus type 1 drug resistance. Proc. Natl. Acad. Sci. USA 2006, 103, 17355–17360. [Google Scholar] [CrossRef] [PubMed]
- Van Westen, G.J.P.; Hendriks, A.; Wegner, J.K.; Ijzerman, A.P.; van Vlijmen, H.W.T.; Bender, A. Significantly improved HIV inhibitor efficacy prediction employing proteochemometric models generated from antivirogram data. PLoS Comput. Biol. 2013, 9, e1002899. [Google Scholar] [CrossRef] [PubMed]
- Tarasova, O.; Filimonov, D.; Poroikov, V. PASS-based approach to predict HIV-1 reverse transcriptase resistance. J. Bioinform. Comput. Biol. 2017, 15, 1650040. [Google Scholar] [CrossRef] [PubMed]
- Tarasova, O.A.; Filimonov, D.A.; Poroikov, V.V. Computational prediction of human immunodeficiency resistance to reverse transcriptase inhibitors. Biomed. Khim. 2017, 63, 457–460. [Google Scholar] [CrossRef] [PubMed]
- Kijak, G.H.; Rubio, A.E.; Pampuro, S.E.; Zala, C.; Cahn, P.; Galli, R.; Montaner, J.S.; Salomón, H. Discrepant results in the interpretation of HIV-1 drug-resistance genotypic data among widely used algorithms. HIV Med. 2003, 4, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Riemenschneider, M.; Heider, D. Current Approaches in Computational Drug Resistance Prediction in HIV. Curr. HIV Res. 2016, 14, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Obermeier, M.; Pironti, A.; Berg, T.; Braun, P.; Däumer, M.; Eberle, J.; Ehret, R.; Kaiser, R.; Kleinkauf, N.; Korn, K.; et al. HIV-GRADE: A publicly available, rules-based drug resistance interpretation algorithm integrating bioinformatic knowledge. Intervirology 2012, 55, 102–107. [Google Scholar] [CrossRef] [PubMed]
- Tural, C.; Ruiz, L.; Holtzer, C.; Schapiro, J.; Viciana, P.; González, J.; Domingo, P.; Boucher, C.; Rey-Joly, C.; Clotet, B. Clinical utility of HIV-1 genotyping and expert advice: The Havana trial. AIDS 2002, 16, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Zazzi, M.; Prosperi, M.; Vicenti, I.; Di Giambenedetto, S.; Callegaro, A.; Bruzzone, B.; Baldanti, F.; Gonnelli, A.; Boeri, E.; Paolini, E.; et al. Rules-based HIV-1 genotypic resistance interpretation systems predict 8 week and 24 week virological antiretroviral treatment outcome and benefit from drug potency weighting. J. Antimicrob. Chemother. 2009, 64, 616–624. [Google Scholar] [CrossRef] [PubMed]
- Pasomsub, E.; Sukasem, C.; Sungkanuparph, S.; Kijsirikul, B.; Chantratita, W. The application of artificial neural networks for phenotypic drug resistance prediction: Evaluation and comparison with other interpretation systems. Jpn. J. Infect. Dis. 2010, 63, 87–94. [Google Scholar] [PubMed]
- Ravela, J.; Betts, B.J.; Brun-Vézinet, F.; Vandamme, A.-M.; Descamps, D.; van Laethem, K.; Smith, K.; Schapiro, J.M.; Winslow, D.L.; Reid, C.; Shafer, R.W.; et al. HIV-1 protease and reverse transcriptase mutation patterns responsible for discordances between genotypic drug resistance interpretation algorithms. J. Acquir. Immune Defic. Syndr. 2003, 33, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Doyon, L.; Payant, C.; Brakier-Gingras, L.; Lamarre, D. Novel Gag-Pol frameshift site in human immunodeficiency virus type 1 variants resistant to protease inhibitors. J. Virol. 1998, 72, 6146–6150. [Google Scholar] [PubMed]
- Tarasova, O.A.; Urusova, A.F.; Filimonov, D.A.; Nicklaus, M.C.; Zakharov, A.V.; Poroikov, V.V. QSAR Modeling Using Large-Scale Databases: Case Study for HIV-1 Reverse Transcriptase Inhibitors. J. Chem. Inf. Model. 2015, 55, 1388–1399. [Google Scholar] [CrossRef] [PubMed]
- Clutter, D.; Sánchez, P.; Rhee, S.-Y.; Shafer, R. Genetic Variability of HIV-1 for Drug Resistance Assay Development. Viruses 2016, 8, 48. [Google Scholar] [CrossRef] [PubMed]
- Bure, D.; Makhdoomi, M.; Lodha, R.; Prakash, S.; Kumar, R.; Parray, H.; Singh, R.; Kabra, S.; Luthra, K. Mutations in the Reverse Transcriptase and Protease Genes of Human Immunodeficiency Virus-1 from Antiretroviral Naïve and Treated Pediatric Patients. Viruses 2015, 7, 590–603. [Google Scholar] [CrossRef] [PubMed]
- Lau, K.A.; Wong, J.J.L. Current Trends of HIV Recombination Worldwide. Infect. Dis. Rep. 2013, 5, e4. [Google Scholar] [CrossRef] [PubMed]
- Junqueira, D.M.; Almeida, S.E. HIV-1 subtype B: Traces of a pandemic. Virology 2016, 495, 173–184. [Google Scholar] [CrossRef] [PubMed]
- Van Westen, G.J.P.; Wegner, J.K.; Geluykens, P.; Kwanten, L.; Vereycken, I.; Peeters, A.; Ijzerman, A.P.; van Vlijmen, H.W.T.; Bender, A. Which compound to select in lead optimization? Prospectively validated proteochemometric models guide preclinical development. PLoS ONE 2011, 6, e27518. [Google Scholar] [CrossRef] [PubMed]
- Tongo, M.; Burgers, W. Challenges in the Design of a T Cell Vaccine in the Context of HIV-1 Diversity. Viruses 2014, 6, 3968–3990. [Google Scholar] [CrossRef] [PubMed]
- Yang, O.O.; Ali, A.; Kasahara, N.; Faure-Kumar, E.; Bae, J.Y.; Picker, L.J.; Park, H. Short Conserved Sequences of HIV-1 Are Highly Immunogenic and Shift Immunodominance. J. Virol. 2015, 89, 1195–1204. [Google Scholar] [CrossRef] [PubMed]
- Hraber, P.; Korber, B.; Wagh, K.; Giorgi, E.; Bhattacharya, T.; Gnanakaran, S.; Lapedes, A.; Learn, G.; Kreider, E.; Li, Y.; et al. Longitudinal Antigenic Sequences and Sites from Intra-Host Evolution (LASSIE) Identifies Immune-Selected HIV Variants. Viruses 2015, 7, 5443–5475. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Verheyen, J.; Rhee, S.-Y.; Voet, A.; Vandamme, A.-M.; Theys, K. Functional conservation of HIV-1 Gag: Implications for rational drug design. Retrovirology 2013, 10, 126. [Google Scholar] [CrossRef] [PubMed]
- López, P.; Rivera-Amill, V.; Rodríguez, N.; Vargas, F.; Yamamura, Y. The Genetic Diversity and Evolution of HIV-1 Subtype B Epidemic in Puerto Rico. Int. J. Environ. Res. Public Health 2015, 13, 55. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhu, J.; Hassink, M.; Jenkins, L.M.M.; Wan, Y.; Appella, D.H.; Xu, J.; Appella, E.; Zhang, X. A novel preventive strategy against HIV-1 infection: Combinatorial use of inhibitors targeting the nucleocapsid and fusion proteins. Emerg. Microbes Infect. 2017, 6, e40. [Google Scholar] [CrossRef] [PubMed]
- Manocheewa, S.; Mittler, J.; Samudrala, R.; Mullins, J. Composite Sequence-Structure Stability Models as Screening Tools for Identifying Vulnerable Targets for HIV Drug and Vaccine Development. Viruses 2015, 7, 5718–5735. [Google Scholar] [CrossRef] [PubMed]



| Name of System/Publication | Data Source * | Algorithm | No of HIV Susceptibility Levels/Another Output | Ref |
|---|---|---|---|---|
| Rega | Proprietary | Rule-based using Boolean expression | 3 levels | [54] |
| HIV Grade | Proprietary | Rule-based | 4 levels | [66] |
| Geno2Pheno | Proprietary | Decision trees; Support vector machines | Quantitative (Prediction of the FR values) | [55,56] |
| Retrogram | Proprietary | Rule-based | 4 levels | [67] |
| Antiretroscan | Proprietary | Rule-based | 5 levels | [68] |
| HIVTrePS | Proprietary | Random Forests | Estimated probability of the treatment success | [57] |
| EuResist | Proprietary | Combined (Bayes network Support Vector Machines, Fuzzy Logic, Case-Based Reasoning and Random Forests) | Estimated probability of the treatment success | [58] |
| The application of artificial neural networks for phenotypic drug resistance prediction: evaluation and comparison with other interpretation systems | Freely available (Stanford HIV resistance database) | Artificial neural networks | 2 levels | [69] |
| Genotypic predictors of human immunodeficiency virus type 1 drug resistance | Freely available (Stanford HIV resistance database) | Decision trees, neural networks, least-squares regression (LSR), SVR, least angle regression (LARS) | 3 levels | [60] |
| Significantly improved HIV inhibitor efficacy prediction employing proteochemometric models generated from Antivirogram data | Proprietary (training), Free available (validation) | Support vector machines | 2 levels of resistance and quantitative prediction of FR value | [61] |
| PASS-based approach to predict HIV-1 reverse transcriptase resistance Computational prediction of human immunodeficiency resistance to reverse transcriptase inhibitors. | Freely available (Stanford HIV resistance database) | PASS-based (modified Bayes) approach/ Set of machine learning methods | Estimated probability of the resistance occurrence/ belonging to a class of resistant variants | [62,63] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tarasova, O.; Poroikov, V. HIV Resistance Prediction to Reverse Transcriptase Inhibitors: Focus on Open Data. Molecules 2018, 23, 956. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules23040956
Tarasova O, Poroikov V. HIV Resistance Prediction to Reverse Transcriptase Inhibitors: Focus on Open Data. Molecules. 2018; 23(4):956. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules23040956
Chicago/Turabian StyleTarasova, Olga, and Vladimir Poroikov. 2018. "HIV Resistance Prediction to Reverse Transcriptase Inhibitors: Focus on Open Data" Molecules 23, no. 4: 956. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules23040956