Cytostatic and Cytotoxic Natural Products against Cancer Cell Models
Abstract
:1. Introduction
2. Results
2.1. Cytostatic/Cytotoxic Evaluation via Cell Proliferation Assay
2.2. Cell Cycle Arrest and Apoptosis
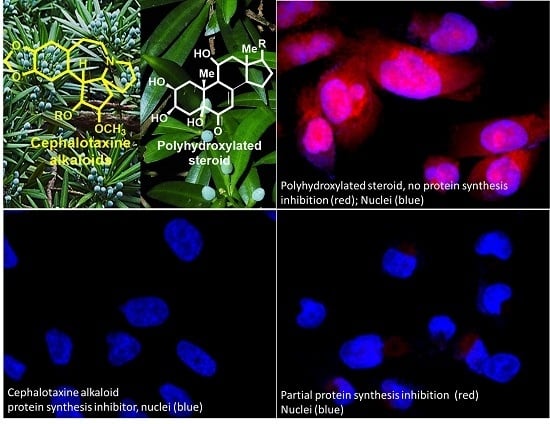
2.3. Protein Synthesis Evaluation
2.4. Probe Synthesis and Evaluation
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Natural Product Compound Library
4.3. CellTiter-Glo Viability Assay (CTG)
4.4. Annexin V-FITC Apoptosis and Cell Cycle
4.5. ApoTox-GloTM Triplex Assay
4.6. Protein Synthesis in Cell-Click Assay
4.7. Probe 9a Evaluation
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs from 1981 to 2014. J. Nat. Prod. 2016, 79, 629–661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gordon, M.; Cragg, D.; Kingston, G.I.; David, M.N. Anticancer Agents from Natural Products, 2nd ed.; CRC Press/Taylor & Francis Group: Boca Raton, FL, USA, 2012. [Google Scholar]
- Harvey, A.L.; Edrada-Ebel, R.; Quinn, R.J. The re-emergence of natural products for drug discovery in the genomics era. Nat. Rev. Drug. Discov. 2015, 14, 111–129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andersson, A.K.; Ma, J.; Wang, J.; Chen, X.; Gedman, A.L.; Dang, J.; Nakitandwe, J.; Holmfeldt, L.; Parker, M.; Easton, J.; et al. Jude Children's Research Hospital–Washington University Pediatric Cancer Genome Project. Nat. Genet. 2015, 47, 330–337. [Google Scholar] [CrossRef] [PubMed]
- Mullighan, C.G.; Miller, C.B.; Radtke, I.; Phillips, L.A.; Dalton, J.; Ma, J.; White, D.; Hughes, T.P.; Le Beau, M.M.; Pui, C.-H.; et al. BCR-ABL1 Lymphoblastic Leukaemia is Characterized by the Deletion of Ikaros. Nature. 2008, 453, 110–114. [Google Scholar] [CrossRef] [PubMed]
- Webersinke, H.R.G. Molecular Pathogenesis of Philadelphia-positive Chronic Myeloid Leukemia–Is It all BCR-ABL? Curr. Cancer Drug Tar. 2011, 11, 3–19. [Google Scholar]
- Wassermann, A.M.; Lounkine, E.; Hoepfner, D.; Le Goff, G.; King, F.J.; Studer, C.; Peltier, J.M.; Grippo, M.L.; Prindle, V.; Tao, J.; et al. Dark Chemical Matter as a Promising Starting point for Drug Lead Discovery. Nat. Chem. Biol. 2015, 11, 958–966. [Google Scholar] [CrossRef]
- Jones, L.H.; Bunnage, M.E. Applications of Chemogenomic Library Screening in Drug Discovery. Nat. Rev. Drug Discov. 2017, 16, 285–296. [Google Scholar] [CrossRef]
- Gezici, S.; Șekeroğlu, N. Current Perspectives in the Application of Medicinal Plants Against Cancer: Novel Therapeutic Agents. Anticancer Agents Med Chem. 2018. [Google Scholar] [CrossRef] [PubMed]
- He, C.Y.; Fu, J.; Shou, J.W.; Zhao, Z.X.; Ren, L.; Wang, Y.; Jiang, J.D. In Vitro Study of the Metabolic Characteristics of Eight Isoquinoline Alkaloids from Natural Plants in Rat Gut Microbiota. Molecules 2017, 22, 932. [Google Scholar] [CrossRef] [PubMed]
- Hadi, V.; Hotard, M.; Ling, T.; Salinas, Y.G.; Palacios, G.; Connelly, M.; Rivas, F. Evaluation of Jatropha Isabelli Natural Products and their Synthetic Analogs as Potential Antimalarial Therapeutic Agents. Eur. J. Med. Chem. 2013, 65, 376–380. [Google Scholar] [CrossRef]
- Mitachi, K.; Salinas, Y.G.; Connelly, M.; Jensen, N.; Ling, T.; Rivas, F. Synthesis and Structure-Activity Relationship of Disubstituted Benzamides as a Novel Class of Antimalarial Agents. Bioorganic Med. Chem. Lett. 2012, 22, 4536–4539. [Google Scholar] [CrossRef] [PubMed]
- Ling, T.; Lang, W.; Feng, X.; Das, S.; Maier, J.; Jeffries, C.; Shelat, A.; Rivas, F. Novel Vitexin-Inspired Scaffold Against Leukemia. Eur. J. Med. Chem. 2018, 146, 501–510. [Google Scholar] [CrossRef]
- Barretina, J.G.; Caponigro, G.; Stransky, N.; Venkatesan, K.; Margolin, A.A.; Kim, S.; Wilson, C.J.; Lehar, J.; Kryukov, G.V.; Sonkin, D.; et al. The Cancer Cell Line Encyclopedia Enables Predictive Modelling of Anticancer Drug Sensitivity. Nature. 2012, 483, 603–607. [Google Scholar] [CrossRef]
- Powell, R.G.; Weisleder, D.; Smith, C.R. Jr.; Rohwedder, W.K. Structures of Harringtonine, Isoharringtonine, and Homoharringtonine. Tetrahedron Lett. 1970, 11, 815–818. [Google Scholar] [CrossRef]
- Takeda, S.; Yajima, N.; Kitazato, K.; Unemi, N. Antitumor Activities of Harringtonine and Homoharringtonine, Cephalotaxus Alkaloids which are Active Principles from Plant by Intraperitoneal and Oral Administration. J. Pharmacobiodyn. 1982, 5, 841–847. [Google Scholar] [CrossRef]
- Abdelkafi, H.; Nay, B. Natural Products from Cephalotaxus sp.: Chemical Diversity and Synthetic Aspects. Nat. Prod. Rep. 2012, 29, 845–869. [Google Scholar] [CrossRef]
- Berman, E. Omacetaxine: The FDA Decision. Clin Adv Hematol Oncol. 2011, 9, 57–58. [Google Scholar] [PubMed]
- Chen, X.; Tang, Y.; Chen, J.; Chen, R.; Gu, L.; Xue, H.; Pan, C.; Tang, J.; Shen, S. Homoharringtonine is a Safe and Effective Substitute for Anthracyclines in Children Younger than 2 Years Old with Acute Myeloid Leukemia. Front Med. 2019. [Google Scholar] [CrossRef] [PubMed]
- Ju, X.; Beaudry, C.M. Total Synthesis of (-)-Cephalotaxine and (-)-Homoharringtonine via Furan Oxidation-Transannular Mannich Cyclization. Angew. Chem. Int. Ed. Engl. 2019. [Google Scholar] [CrossRef]
- Min, X.; Na, Z.; Yanan, L.; Chunrui, L. De Novo Acute Megakaryoblastic Leukemia with p210 BCR/ABL and t(1;16) Translocation but not t(9;22) Ph Chromosome. J. Hematol. Oncol. 2011, 4, 45. [Google Scholar] [CrossRef]
- Jiang, T.L.; Liu, R.H.; Salmon, S.E. Comparative in vitro Antitumor Activity of Homoharringtonine and Harringtonine against Clonogenic Human Tumor Cells. Invest. New. Drugs. 1983, 1, 21–25. [Google Scholar] [CrossRef] [PubMed]
- Hwang, B.Y.; Su, B.N.; Chai, H.; Mi, Q.; Kardono, L.B.; Afriastini, J.J.; Riswan, S.; Santarsiero, B.D.; Mesecar, A.D.; Wild, R.; et al. Silvestrol and Episilvestrol, Potential Anticancer Rocaglate Derivatives from Aglaia Silvestris. J. Org. Chem. 2004, 69, 3350–3358. [Google Scholar] [CrossRef] [PubMed]
- Rodrigo, C.M.; Cencic, R.; Roche, S.P.; Pelletier, J.; Porco, J.A. Synthesis of Rocaglamide Hydroxamates and Related Compounds as Eukaryotic Translation Inhibitors: Synthetic and Biological Studies. J. Med. Chem. 2012, 55, 558–562. [Google Scholar] [CrossRef]
- Kogure, T.; Kinghorn, A.D.; Yan, I.; Bolon, B.; Lucas, D.M.; Grever, M.R.; Patel, T. Therapeutic Potential of the Translation Inhibitor Silvestrol in Hepatocellular Cancer. PLoS ONE. 2013, 8, e76136. [Google Scholar] [CrossRef] [PubMed]
- Lucas, D.M.; Edwards, R.B.; Lozanski, G.; West, D.A.; Shin, J.D.; Vargo, M.A.; Davis, M.E.; Rozewski, D.M.; Johnson, A.J.; Su, B.N.; et al. The Novel Plant-derived agent Silvestrol has B-Cell Selective Activity in Chronic Lymphocytic Leukemia and Acute Lymphoblastic Leukemia in Vitro and in Vivo. Blood. 2009, 113, 4656–4666. [Google Scholar] [CrossRef]
- Kim, U.; Shu, C.W.; Dane, K.Y.; Daugherty, P.S.; Wang, J.Y.; Soh, H.T. Selection of Mammalian Cells Based on their Cell-cycle Phase using Dielectrophoresis. Proc. Natl. Acad. Sci. USA 2007, 104, 20708–20712. [Google Scholar] [CrossRef]
- Crowley, L.C.; Marfell, B.J.; Scott, A.P.; Waterhouse, N.J. Quantitation of Apoptosis and Necrosis by Annexin V Binding, Propidium Iodide Uptake, and Flow Cytometry. Cold Spring Harb. Protoc. 2016. [Google Scholar] [CrossRef]
- Shalini, S.; Dorstyn, L.; Dawar, S.; Kumar, S. Old, New and Emerging Functions of Caspases. Cell Death Differ. 2015, 22, 526–539. [Google Scholar] [CrossRef]
- Kastan, M.B.; Bartek, J. Cell-cycle Checkpoints and Cancer. Nature. 2004, 432, 316–323. [Google Scholar] [CrossRef] [PubMed]
- Riccardi, C.; Nicoletti, I. Analysis of Apoptosis by Propidium Iodide Staining and Flow Cytometry. Nat. Protoc. 2006, 1, 1458–1461. [Google Scholar] [CrossRef]
- Joazeiro, C.A.P. Mechanisms and Functions of Ribosome-Associated Protein Quality Control. Nat. Rev. Mol. Cell. Biol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Xu, Y.; Stoleru, D.; Salic, A. Imaging Protein Synthesis in Cells and Tissues with an Alkyne Analog of Puromycin. Proc. Natl. Acad. Sci. USA 2012, 109, 413–418. [Google Scholar] [CrossRef]
- Cuyàs, E.; Martin-Castillo, B.; Corominas-Faja, B.; Massaguer, A.; Bosch-Barrera, J.; Menendez, J.A. Anti-Protozoal and Anti-Bacterial Antibiotics that Inhibit Protein Synthesis Kill Cancer Subtypes Enriched for Stem Cell-like Properties. Cell Cycle. 2015, 14, 3527–3532. [Google Scholar] [CrossRef] [PubMed]
- Burger, K.; Mühl, B.; Harasim, T.; Rohrmoser, M.; Malamoussi, A.; Orban, M.; Kellner, M.; Gruber-Eber, A.; Kremmer, E.; Hölzel, M.; et al. Chemotherapeutic Drugs Inhibit Ribosome Biogenesis at Various Levels. J. Biol. Chem. 2010, 285, 12416–12425. [Google Scholar] [CrossRef] [Green Version]
- Zhu, H.; Fan, J.; Du, J.; Peng, X. Fluorescent Probes for Sensing and Imaging within Specific Cellular Organelles. Acc. Chem. Res. 2016, 49, 2115–2126. [Google Scholar] [CrossRef]
- Bhute, V.J.; Ma, Y.; Bao, X.; Palecek, S.P. The Poly (ADP-Ribose) Polymerase Inhibitor Veliparib and Radiation Cause Significant Cell Line Dependent Metabolic Changes in Breast Cancer Cells. Sci. Rep. 2016, 6, 36061. [Google Scholar] [CrossRef] [Green Version]
- Smith, C.R.; Powell, R.G.; Suffness, M. Development of Homoharringtonine. J. Clin. Oncol. 1986, 4, 1283. [Google Scholar] [CrossRef]
- Zhong, S.B.; Liu, W.C.; Li, R.L.; Ling, Y.Z.; Li, C.H.; Tu, G.Z.; Ma, L.B.; Hong, S.L. Studies on Semi-synthesis of Cephalotaxine Esters and Correlation of their Structures with Antitumor Activity. Yao Xue Xue Bao. 1994, 29, 33–38. [Google Scholar] [PubMed]
- Fresno, M.; Jiménez, A.; Vázquez, D. Inhibition of Translation in Eukaryotic Systems by Harringtonine. Eur. J. Biochem. 1977, 72, 323–330. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.E.; Song, Y.J. Anti-varicella-zoster Virus Activity of Cephalotaxine Esters in Vitro. J. Microbiol. 2019, 57, 74–79. [Google Scholar] [CrossRef]
- Gürel, G.; Blaha, G.; Moore, P.B.; Steitz, T.A. U2504 Determines the Species Specificity of the A-site Cleft Antibiotics: The Structures of Tiamulin, Homoharringtonine, and Bruceantin Bound to the Ribosome. J. Mol. Biol. 2009, 389, 46–56. [Google Scholar] [CrossRef] [PubMed]
- Hay, R.J.; Caputo, J.L.; Macy, M.L. ATCC Quality Control Methods for Cell Lines, 2nd ed.; ATCC: Manassas, VA, USA, 1992. [Google Scholar]
Sample Availability: Samples of the compounds are available from the authors. |









| SUP-B15 (EC50 µM) | KOPN-8 (EC50 µM) | NALM-06 (EC50 µM) | UoC-B1 (EC50 µM) | BJ (EC50 µM) | PBMC (EC50 µM) | TI (PBMC/SUP-B15) | |
|---|---|---|---|---|---|---|---|
| 1 | 0.1201 | 0.41 ± 0.05 | 0.41 ± 0.15 | 0.0401 ± 0.01 | >38.8788 | >38.8788 | >316 |
| 4 | 38 ± 6.1 | 32.1 ± 3.5 | 18.3 ± 2.5 | >23.3766 | >46.7532 | >86.7532 | >2.25 |
| 5 | 29 ± 3.7 | 32 ± 7.6 | 8.3832 ± 1.8 | 17.28 ± 3.8 | >43.2900 | >43.2900 | >1.48 |
| 6 | 9.83 ± 0.5 | 0.1602 ± 0.05 | 18.45 ± 3.1 | >21.6450 | >43.2900 | >43.2900 | >4.3 |
| 7 | 6.05 ± 0.8 | 4.0732 ± 0.45 | 3.7229 ± 0.5 | 2.88 ± 1.1 | >51.9481 | 28.0668 | 4.6 |
| 8 | 2.77 ± 0.36 | 0.8208 ± 0.22 | 1.9 ± 0.2 | 1.96 ± 0.28 | 29.8382 | 7.8914 | 2.8 |
| 9 | 10.43 ± 0.75 | 23.12 ± 5.2 | 9.5 ± 2.7 | 17.6 ± 5.8 | >43.2900 | >83 | >7.7 |
| 10 | 0.045 ± 0.005 | 0.0799 ± 0.01 | 0.0322 ± 0.01 | 0.0129 ± 0.002 | >43.2900 | 6.3499 | 140 |
| 11 | 29 ± 6 | >43.2900 | >43.2900 | >21.6450 | >43.2900 | >43.2900 | >1.48 |
| 12 | 20.1 ± 3.2 | >43.2900 | >43.2900 | >21.6450 | >43.2900 | >43.2900 | >2.15 |
| 13 | 9.5 ± 2.1 | >43.2900 | 0.518 ± 0.15 | >21.6450 | >43.2900 | >43.2900 | >4.5 |
| 14 | 17.2 ± 4.5 | >43.2900 | >43.2900 | >21.6450 | >43.2900 | >43.2900 | >2.5 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ling, T.; Lang, W.H.; Maier, J.; Quintana Centurion, M.; Rivas, F. Cytostatic and Cytotoxic Natural Products against Cancer Cell Models. Molecules 2019, 24, 2012. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules24102012
Ling T, Lang WH, Maier J, Quintana Centurion M, Rivas F. Cytostatic and Cytotoxic Natural Products against Cancer Cell Models. Molecules. 2019; 24(10):2012. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules24102012
Chicago/Turabian StyleLing, Taotao, Walter H. Lang, Julie Maier, Marizza Quintana Centurion, and Fatima Rivas. 2019. "Cytostatic and Cytotoxic Natural Products against Cancer Cell Models" Molecules 24, no. 10: 2012. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules24102012