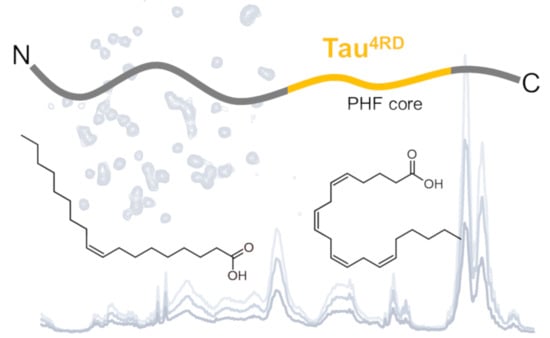
Unsaturated Fatty Acid-Induced Conformational Transitions and Aggregation of the Repeat Domain of Tau
Abstract
:1. Introduction
2. Results
2.1. Aggregation State of Oleic and Arachidonic Acids in an Aqueous Solution
2.2. Association of Tau4RD with Fatty Acids
2.3. Site-Resolved Investigation of Fatty Acid-Induced Perturbations of Tau4RD
2.4. Time-Dependent Tau4RD Monomer Depletion and Translational Diffusion
2.5. FA-Induced Protein Secondary Structure Perturbations
2.6. FA-Induced Fibril Formation
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Recombinant Protein Expression and Purification
4.3. Fatty Acid Samples Preparation
4.4. NMR Spectroscopy
4.5. Fluorescence Spectroscopy
4.6. Far-UV Circular Dichroism (CD) Spectroscopy
4.7. Thioflavin-T Aggregation Assay
4.8. Transmission Electron Microscopy (TEM)
4.9. Turbidimetry
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Weingarten, M.D.; Lockwood, A.H.; Hwo, S.Y.; Kirschner, M.W. A protein factor essential for microtubule assembly. Proc. Natl. Acad. Sci. USA 1975, 72, 1858–1862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drubin, D.G.; Kirschner, M.W. Tau protein function in living cells. J. Cell Biol. 1986, 103, 2739–2746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morris, M.; Maeda, S.; Vossel, K.; Mucke, L. The Many Faces of Tau. Neuron 2011, 70, 410–426. [Google Scholar] [CrossRef] [Green Version]
- Tapia-Rojas, C.; Cabezas-Opazo, F.; Deaton, C.A.; Vergara, E.H.; Johnson, G.V.W.; Quintanilla, R.A. It’s all about tau. Prog. Neurobiol. 2019, 175, 54–76. [Google Scholar] [CrossRef] [PubMed]
- Schweers, O.; Schönbrunn-Hanebeck, E.; Marx, A.; Mandelkow, E. Structural studies of tau protein and Alzheimer paired helical filaments show no evidence for beta-structure. J. Biol. Chem. 1994, 269, 24290–24297. [Google Scholar]
- Martin, L.; Latypova, X.; Terro, F. Post-translational modifications of tau protein: Implications for Alzheimer’s disease. Neurochem. Int. 2011, 58, 458–471. [Google Scholar] [CrossRef] [PubMed]
- Goedert, M.; Spillantini, M.G.; Potier, M.C.; Ulrich, J.; Crowther, R.A. Cloning and sequencing of the cDNA encoding an isoform of microtubule-associated protein tau containing four tandem repeats: Differential expression of tau protein mRNAs in human brain. EMBO J. 1989, 8, 393–399. [Google Scholar] [CrossRef]
- Wang, Y.; Mandelkow, E. Tau in physiology and pathology. Nat. Rev. Neurosci. 2016, 17, 22–35. [Google Scholar] [CrossRef]
- Von Bergen, M.; Friedhoff, P.; Biernat, J.; Heberle, J.; Mandelkow, E.-M.; Mandelkow, E. Assembly of tau protein into Alzheimer paired helical filaments depends on a local sequence motif (306VQIVYK311) forming beta structure. Proc. Natl. Acad. Sci. USA 2000, 97, 5129–5134. [Google Scholar] [CrossRef] [Green Version]
- Mukrasch, M.D.; Bibow, S.; Korukottu, J.; Jeganathan, S.; Biernat, J.; Griesinger, C.; Mandelkow, E.; Zweckstetter, M. Structural Polymorphism of 441-Residue Tau at Single Residue Resolution. PLoS Biol. 2009, 7, e1000034. [Google Scholar] [CrossRef]
- Bielska, A.A.; Zondlo, N.J. Hyperphosphorylation of Tau Induces Local Polyproline II Helix. Biochemistry 2006, 45, 5527–5537. [Google Scholar] [CrossRef]
- Schwalbe, M.; Kadavath, H.; Biernat, J.; Ozenne, V.; Blackledge, M.; Mandelkow, E.; Zweckstetter, M. Structural Impact of Tau Phosphorylation at Threonine 231. Structure 2015, 23, 1448–1458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kadavath, H.; Jaremko, M.; Jaremko, Ł.; Biernat, J.; Mandelkow, E.; Zweckstetter, M. Folding of the Tau Protein on Microtubules. Angew. Chem. Int. Ed. 2015, 54, 10347–10351. [Google Scholar] [CrossRef] [PubMed]
- Kellogg, E.H.; Hejab, N.M.A.; Poepsel, S.; Downing, K.H.; DiMaio, F.; Nogales, E. Near-atomic model of microtubule-tau interactions. Science 2018, 360, 1242–1246. [Google Scholar] [CrossRef] [Green Version]
- Grundke-Iqbal, I.; Iqbal, K.; Quinlan, M.; Tung, Y.C.; Zaidi, M.S.; Wisniewski, H.M. Microtubule-associated protein tau. A component of Alzheimer paired helical filaments. J. Biol. Chem. 1986, 261, 6084–6089. [Google Scholar] [PubMed]
- Kosik, K.S.; Joachim, C.L.; Selkoe, D.J. Microtubule-associated protein tau (tau) is a major antigenic component of paired helical filaments in Alzheimer disease. Proc. Natl. Acad. Sci. USA 1986, 83, 4044–4048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, V.M.-Y.; Goedert, M.; Trojanowski, J.Q. Neurodegenerative Tauopathies. Annu. Rev. Neurosci. 2001, 24, 1121–1159. [Google Scholar] [CrossRef]
- Fitzpatrick, A.W.P.; Falcon, B.; He, S.; Murzin, A.G.; Murshudov, G.; Garringer, H.J.; Crowther, R.A.; Ghetti, B.; Goedert, M.; Scheres, S.H.W. Cryo-EM structures of tau filaments from Alzheimer’s disease. Nature 2017, 547, 185–190. [Google Scholar] [CrossRef] [Green Version]
- Falcon, B.; Zhang, W.; Murzin, A.G.; Murshudov, G.; Garringer, H.J.; Vidal, R.; Crowther, R.A.; Ghetti, B.; Scheres, S.H.W.; Goedert, M. Structures of filaments from Pick’s disease reveal a novel tau protein fold. Nature 2018, 561, 137–140. [Google Scholar] [CrossRef]
- Jeganathan, S.; von Bergen, M.; Brutlach, H.; Steinhoff, H.-J.; Mandelkow, E. Global hairpin folding of tau in solution. Biochemistry 2006, 45, 2283–2293. [Google Scholar] [CrossRef]
- Novak, P.; Cehlar, O.; Skrabana, R.; Novak, M. Tau Conformation as a Target for Disease-Modifying Therapy: The Role of Truncation. J. Alzheimers Dis. 2018, 64, S535–S546. [Google Scholar] [CrossRef]
- Mandelkow, E.-M.; Mandelkow, E. Biochemistry and Cell Biology of Tau Protein in Neurofibrillary Degeneration. Cold Spring Harb. Perspect. Med. 2012, 2, a006247. [Google Scholar] [CrossRef]
- Goedert, M.; Jakes, R.; Spillantini, M.G.; Hasegawa, M.; Smith, M.J.; Crowther, R.A. Assembly of microtubule-associated protein tau into Alzheimer-like filaments induced by sulphated glycosaminoglycans. Nature 1996, 383, 550–553. [Google Scholar] [CrossRef]
- Kampers, T.; Friedhoff, P.; Biernat, J.; Mandelkow, E.M.; Mandelkow, E. RNA stimulates aggregation of microtubule-associated protein tau into Alzheimer-like paired helical filaments. FEBS Lett. 1996, 399, 344–349. [Google Scholar] [CrossRef]
- Wilson, D.M.; Binder, L.I. Free fatty acids stimulate the polymerization of tau and amyloid beta peptides. In vitro evidence for a common effector of pathogenesis in Alzheimer’s disease. Am. J. Pathol. 1997, 150, 2181–2195. [Google Scholar]
- Brandt, R. Interaction of tau with the neural plasma membrane mediated by tau’s amino-terminal projection domain. J. Cell Biol. 1995, 131, 1327–1340. [Google Scholar] [CrossRef]
- Farah, C.A.; Perreault, S.; Liazoghli, D.; Desjardins, M.; Anton, A.; Lauzon, M.; Paiement, J.; Leclerc, N. Tau interacts with Golgi membranes and mediates their association with microtubules. Cell Motil. Cytoskelet. 2006, 63, 710–724. [Google Scholar] [CrossRef] [PubMed]
- Mutreja, Y.; Gamblin, T.C. Optimization of in vitro conditions to study the arachidonic acid induction of 4R isoforms of the microtubule-associated protein tau. Methods Cell Biol. 2017, 141, 65–88. [Google Scholar] [CrossRef]
- Patil, S.; Chan, C. Palmitic and stearic fatty acids induce Alzheimer-like hyperphosphorylation of tau in primary rat cortical neurons. Neurosci. Lett. 2005, 384, 288–293. [Google Scholar] [CrossRef] [PubMed]
- Kitajka, K.; Puskás, L.G.; Zvara, A.; Hackler, L.; Barceló-Coblijn, G.; Yeo, Y.K.; Farkas, T. The role of n-3 polyunsaturated fatty acids in brain: Modulation of rat brain gene expression by dietary n-3 fatty acids. Proc. Natl. Acad. Sci. USA 2002, 99, 2619–2624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Broersen, K.; van den Brink, D.; Fraser, G.; Goedert, M.; Davletov, B. Alpha-synuclein adopts an alpha-helical conformation in the presence of polyunsaturated fatty acids to hinder micelle formation. Biochemistry 2006, 45, 15610–15616. [Google Scholar] [CrossRef]
- De Franceschi, G.; Frare, E.; Pivato, M.; Relini, A.; Penco, A.; Greggio, E.; Bubacco, L.; Fontana, A.; de Laureto, P.P. Structural and morphological characterization of aggregated species of α-synuclein induced by docosahexaenoic acid. J. Biol. Chem. 2011, 286, 22262–22274. [Google Scholar] [CrossRef] [Green Version]
- Gray, E.G.; Paula-Barbosa, M.; Roher, A. Alzheimer’s disease: Paired helical filaments and cytomembranes. Neuropathol. Appl. Neurobiol. 1987, 13, 91–110. [Google Scholar] [CrossRef]
- Goux, W.J.; Rodriguez, S.; Sparkman, D.R. Analysis of the core components of Alzheimer paired helical filaments. A gas chromatography/mass spectrometry characterization of fatty acids, carbohydrates and long-chain bases. FEBS Lett. 1995, 366, 81–85. [Google Scholar] [CrossRef] [Green Version]
- Sparr, E.; Linse, S. Lipid-protein interactions in amyloid formation. Biochim. Biophys. Acta Proteins Proteom. 2019, 1867, 455–457. [Google Scholar] [CrossRef]
- Ait-Bouziad, N.; Lv, G.; Mahul-Mellier, A.-L.; Xiao, S.; Zorludemir, G.; Eliezer, D.; Walz, T.; Lashuel, H.A. Discovery and characterization of stable and toxic Tau/phospholipid oligomeric complexes. Nat. Commun. 2017, 8, 1678. [Google Scholar] [CrossRef] [Green Version]
- Deryusheva, E.; Nemashkalova, E.; Galloux, M.; Richard, C.; Eléouët, J.; Kovacs, D.; Belle, K.; Tompa, P.; Uversky, V.; Permyakov, S. Does Intrinsic Disorder in Proteins Favor Their Interaction with Lipids? Proteomics 2019, 19, 1800098. [Google Scholar] [CrossRef]
- Killinger, B.A.; Melki, R.; Brundin, P.; Kordower, J.H. Endogenous alpha-synuclein monomers, oligomers and resulting pathology: Let’s talk about the lipids in the room. Npj Parkinsons Dis. 2019, 5, 23. [Google Scholar] [CrossRef] [Green Version]
- Cistola, D.P.; Hamilton, J.A.; Jackson, D.; Small, D.M. Ionization and phase behavior of fatty acids in water: Application of the Gibbs phase rule. Biochemistry 1988, 27, 1881–1888. [Google Scholar] [CrossRef]
- Serth, J.; Lautwein, A.; Frech, M.; Wittinghofer, A.; Pingoud, A. The inhibition of the GTPase activating protein-Ha-ras interaction by acidic lipids is due to physical association of the C-terminal domain of the GTPase activating protein with micellar structures. EMBO J. 1991, 10, 1325–1330. [Google Scholar] [CrossRef]
- Lashuel, H.A.; Overk, C.R.; Oueslati, A.; Masliah, E. The many faces of α-synuclein: From structure and toxicity to therapeutic target. Nat. Rev. Neurosci. 2013, 14, 38–48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alza, N.P.; Iglesias González, P.A.; Conde, M.A.; Uranga, R.M.; Salvador, G.A. Lipids at the Crossroad of α-Synuclein Function and Dysfunction: Biological and Pathological Implications. Front. Cell. Neurosci. 2019, 13, 175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munari, F.; D’Onofrio, M.; Assfalg, M. Solution NMR insights into dynamic supramolecular assemblies of disordered amyloidogenic proteins. Arch. Biochem. Biophys. 2020, 683, 108304. [Google Scholar] [CrossRef] [PubMed]
- Brutscher, B.; Felli, I.C.; Gil-Caballero, S.; Hošek, T.; Kümmerle, R.; Piai, A.; Pierattelli, R.; Sólyom, Z. NMR Methods for the Study of Instrinsically Disordered Proteins Structure, Dynamics, and Interactions: General Overview and Practical Guidelines. In Intrinsically Disordered Proteins Studied by NMR Spectroscopy; Felli, I.C., Pierattelli, R., Eds.; Springer International Publishing: Cham, Switzerland, 2015; Volume 870, pp. 49–122. ISBN 978-3-319-20163-4. [Google Scholar]
- Anthis, N.J.; Clore, G.M. Visualizing transient dark states by NMR spectroscopy. Quart. Rev. Biophys. 2015, 48, 35–116. [Google Scholar] [CrossRef]
- Kleckner, I.R.; Foster, M.P. An introduction to NMR-based approaches for measuring protein dynamics. Biochim. Biophys. Acta 2011, 1814, 942–968. [Google Scholar] [CrossRef] [Green Version]
- Schweers, O.; Mandelkow, E.M.; Biernat, J.; Mandelkow, E. Oxidation of cysteine-322 in the repeat domain of microtubule-associated protein tau controls the in vitro assembly of paired helical filaments. Proc. Natl. Acad. Sci. USA 1995, 92, 8463–8467. [Google Scholar] [CrossRef] [Green Version]
- Karikari, T.K.; Turner, A.; Stass, R.; Lee, L.C.Y.; Wilson, B.; Nagel, D.A.; Hill, E.J.; Moffat, K.G. Expression and purification of tau protein and its frontotemporal dementia variants using a cleavable histidine tag. Protein Expr. Purif. 2017, 130, 44–54. [Google Scholar] [CrossRef] [Green Version]
- Barré, P.; Eliezer, D. Structural transitions in tau k18 on micelle binding suggest a hierarchy in the efficacy of individual microtubule-binding repeats in filament nucleation: Hierarchy in the Efficacy of Tau Repeats in Filament Nucleatior. Protein Sci. 2013, 22, 1037–1048. [Google Scholar] [CrossRef] [Green Version]
- Iljina, M.; Tosatto, L.; Choi, M.L.; Sang, J.C.; Ye, Y.; Hughes, C.D.; Bryant, C.E.; Gandhi, S.; Klenerman, D. Arachidonic acid mediates the formation of abundant alpha-helical multimers of alpha-synuclein. Sci. Rep. 2016, 6, 33928. [Google Scholar] [CrossRef] [Green Version]
- King, M.E.; Ahuja, V.; Binder, L.I.; Kuret, J. Ligand-Dependent Tau Filament Formation: Implications for Alzheimer’s Disease Progression. Biochemistry 1999, 38, 14851–14859. [Google Scholar] [CrossRef]
- Zhang, W.; Falcon, B.; Murzin, A.G.; Fan, J.; Crowther, R.A.; Goedert, M.; Scheres, S.H. Heparin-induced tau filaments are polymorphic and differ from those in Alzheimer’s and Pick’s diseases. eLife 2019, 8, e43584. [Google Scholar] [CrossRef]
- Elbaum-Garfinkle, S.; Ramlall, T.; Rhoades, E. The role of the lipid bilayer in tau aggregation. Biophys. J. 2010, 98, 2722–2730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chirita, C.N.; Necula, M.; Kuret, J. Anionic Micelles and Vesicles Induce Tau Fibrillization in Vitro. J. Biol. Chem. 2003, 278, 25644–25650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Georgieva, E.R.; Xiao, S.; Borbat, P.P.; Freed, J.H.; Eliezer, D. Tau binds to lipid membrane surfaces via short amphipathic helices located in its microtubule-binding repeats. Biophys. J. 2014, 107, 1441–1452. [Google Scholar] [CrossRef] [Green Version]
- Jones, E.M.; Dubey, M.; Camp, P.J.; Vernon, B.C.; Biernat, J.; Mandelkow, E.; Majewski, J.; Chi, E.Y. Interaction of tau protein with model lipid membranes induces tau structural compaction and membrane disruption. Biochemistry 2012, 51, 2539–2550. [Google Scholar] [CrossRef] [Green Version]
- Micsonai, A.; Wien, F.; Bulyáki, É.; Kun, J.; Moussong, É.; Lee, Y.-H.; Goto, Y.; Réfrégiers, M.; Kardos, J. BeStSel: A web server for accurate protein secondary structure prediction and fold recognition from the circular dichroism spectra. Nucleic Acids Res. 2018, 46, W315–W322. [Google Scholar] [CrossRef] [PubMed]
- Miles, A.J.; Wallace, B.A. Circular dichroism spectroscopy of membrane proteins. Chem. Soc. Rev. 2016, 45, 4859–4872. [Google Scholar] [CrossRef] [Green Version]
- Lipton, P. Ischemic Cell Death in Brain Neurons. Physiol. Rev. 1999, 79, 1431–1568. [Google Scholar] [CrossRef]
- Munari, F.; Barracchia, C.G.; Franchin, C.; Parolini, F.; Capaldi, S.; Romeo, A.; Bubacco, L.; Assfalg, M.; Arrigoni, G.; D’Onofrio, M. Semisynthetic and Enzyme-Mediated Conjugate Preparations Illuminate the Ubiquitination-Dependent Aggregation of Tau Protein. Angew. Chem. Int. Ed. 2020, 59, 6607–6611. [Google Scholar] [CrossRef]
- Plotegher, N.; Kumar, D.; Tessari, I.; Brucale, M.; Munari, F.; Tosatto, L.; Belluzzi, E.; Greggio, E.; Bisaglia, M.; Capaldi, S.; et al. The chaperone-like protein 14-3-3η interacts with human α-synuclein aggregation intermediates rerouting the amyloidogenic pathway and reducing α-synuclein cellular toxicity. Hum. Mol. Genet. 2014, 23, 5615–5629. [Google Scholar] [CrossRef] [Green Version]
- Tira, R.; De Cecco, E.; Rigamonti, V.; Santambrogio, C.; Barracchia, C.G.; Munari, F.; Romeo, A.; Legname, G.; Prosperi, D.; Grandori, R.; et al. Dynamic molecular exchange and conformational transitions of alpha-synuclein at the nano-bio interface. Int. J. Biol. Macromol. 2020, 154, 206–216. [Google Scholar] [CrossRef] [PubMed]
- Zanzoni, S.; Pagano, K.; D’Onofrio, M.; Assfalg, M.; Ciambellotti, S.; Bernacchioni, C.; Turano, P.; Aime, S.; Ragona, L.; Molinari, H. Unsaturated Long-Chain Fatty Acids Are Preferred Ferritin Ligands That Enhance Iron Biomineralization. Chem. Eur. J. 2017, 23, 9879–9887. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.; Tonelli, M.; Markley, J.L. NMRFAM-SPARKY: Enhanced software for biomolecular NMR spectroscopy. Bioinformatics 2015, 31, 1325–1327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sibille, N.; Huvent, I.; Fauquant, C.; Verdegem, D.; Amniai, L.; Leroy, A.; Wieruszeski, J.-M.; Lippens, G.; Landrieu, I. Structural characterization by nuclear magnetic resonance of the impact of phosphorylation in the proline-rich region of the disordered Tau protein. Proteins 2012, 80, 454–462. [Google Scholar] [CrossRef] [PubMed]
- Bermel, W.; Bertini, I.; Duma, L.; Felli, I.C.; Emsley, L.; Pierattelli, R.; Vasos, P.R. Complete Assignment of Heteronuclear Protein Resonances by Protonless NMR Spectroscopy. Angew. Chem. Int. Ed. 2005, 44, 3089–3092. [Google Scholar] [CrossRef] [PubMed]
- Santambrogio, C.; Favretto, F.; D’Onofrio, M.; Assfalg, M.; Grandori, R.; Molinari, H. Mass spectrometry and NMR analysis of ligand binding by human liver fatty acid binding protein: ESI-MS and NMR of hL-FABP complexes. J. Mass Spectrom. 2013, 48, 895–903. [Google Scholar] [CrossRef]
- D’Onofrio, M.; Barracchia, C.G.; Bortot, A.; Munari, F.; Zanzoni, S.; Assfalg, M. Molecular differences between human liver fatty acid binding protein and its T94A variant in their unbound and lipid-bound states. Biochim. Biophys. Acta Proteins Proteom. 2017, 1865, 1152–1159. [Google Scholar] [CrossRef] [PubMed]
- D’Onofrio, M.; Gianolio, E.; Ceccon, A.; Arena, F.; Zanzoni, S.; Fushman, D.; Aime, S.; Molinari, H.; Assfalg, M. High relaxivity supramolecular adducts between human-liver fatty-acid-binding protein and amphiphilic Gd(III) complexes: Structural basis for the design of intracellular targeting MRI probes. Chem. Eur. J. 2012, 18, 9919–9928. [Google Scholar] [CrossRef]
- Favretto, F.; Santambrogio, C.; D’Onofrio, M.; Molinari, H.; Grandori, R.; Assfalg, M. Bile salt recognition by human liver fatty acid binding protein. FEBS J. 2015, 282, 1271–1288. [Google Scholar] [CrossRef] [Green Version]
Sample Availability: Samples of Tau4RD are available from the authors upon request. |










© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barracchia, C.G.; Tira, R.; Parolini, F.; Munari, F.; Bubacco, L.; Spyroulias, G.A.; D’Onofrio, M.; Assfalg, M. Unsaturated Fatty Acid-Induced Conformational Transitions and Aggregation of the Repeat Domain of Tau. Molecules 2020, 25, 2716. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules25112716
Barracchia CG, Tira R, Parolini F, Munari F, Bubacco L, Spyroulias GA, D’Onofrio M, Assfalg M. Unsaturated Fatty Acid-Induced Conformational Transitions and Aggregation of the Repeat Domain of Tau. Molecules. 2020; 25(11):2716. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules25112716
Chicago/Turabian StyleBarracchia, Carlo Giorgio, Roberto Tira, Francesca Parolini, Francesca Munari, Luigi Bubacco, Georgios A. Spyroulias, Mariapina D’Onofrio, and Michael Assfalg. 2020. "Unsaturated Fatty Acid-Induced Conformational Transitions and Aggregation of the Repeat Domain of Tau" Molecules 25, no. 11: 2716. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules25112716