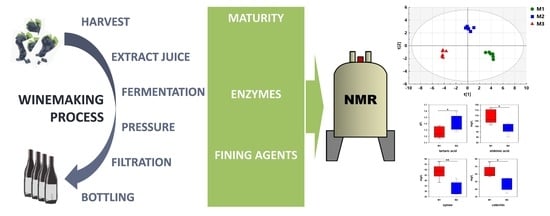
1H-NMR Metabolomics as a Tool for Winemaking Monitoring
Abstract
:1. Introduction
2. Results and Discussion
2.1. 1H-NMR Analysis of Wine
2.2. Maturity Stages
2.3. Enzyme Treatments
2.4. Fining Treatments
3. Materials and Methods
3.1. Wine Samples
3.2. Winemaking
3.3. Sample Preparation
3.4. NMR Analysis
3.5. Multivariate Data Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Cevallos-Cevallos, J.M.; Reyes-De-Corcuera, J.I.; Etxeberria, E.; Danyluk, M.D.; Rodrick, G.E. Metabolomic analysis in food science: A review. Trends Food Sci. Technol. 2009, 20, 557–566. [Google Scholar] [CrossRef]
- Kim, S.; Kim, J.; Yun, E.J.; Kim, K.H. Food metabolomics: From farm to human. Curr. Opin. Biotechnol. 2016, 37, 16–23. [Google Scholar] [CrossRef]
- Valls Fonayet, J.; Loupit, G.; Richard, T. Chapter Ten—MS- and NMR-metabolomic tools for the discrimination of wines: Applications for authenticity. Adv. Bot. Res. 2021, 98, 297–357. [Google Scholar]
- Viskić, M.; Bandić, L.M.; Korenika, A.-M.J.; Jeromel, A. NMR in the service of wine differentiation. Foods 2021, 10, 120. [Google Scholar] [CrossRef] [PubMed]
- Solovyev, P.A.; Fauhl-Hassek, C.; Riedl, J.; Esslinger, S.; Bontempo, L.; Camin, F. NMR spectroscopy in wine authentication: An official control perspective. Compr. Rev. Food Sci. Food Saf. 2021, 20, 2040–2062. [Google Scholar] [CrossRef] [PubMed]
- Sobolev, A.P.; Thomas, F.; Donarski, J.; Ingallina, C.; Circi, S.; Cesare Marincola, F.; Capitani, D.; Mannina, L. Use of NMR applications to tackle future food fraud issues. Trends Food Sci. Technol. 2019, 91, 347–353. [Google Scholar] [CrossRef] [Green Version]
- Tabago, M.K.A.G.; Calingacion, M.N.; Garcia, J. Recent advances in NMR-based metabolomics of alcoholic beverages. Food Chem. Mol. Sci. 2021, 2, 100009. [Google Scholar] [CrossRef]
- Gougeon, L.; Da Costa, G.; Le Mao, I.; Ma, W.; Teissedre, P.-L.; Guyon, F.; Richard, T. Wine analysis and authenticity using 1H-NMR metabolomics data: Application to Chinese wines. Food Anal. Methods 2018, 11, 3425–3434. [Google Scholar] [CrossRef]
- Gougeon, L.; da Costa, G.; Guyon, F.; Richard, T. 1H NMR metabolomics applied to Bordeaux red wines. Food Chem. 2019, 301, 125257. [Google Scholar] [CrossRef] [PubMed]
- Godelmann, R.; Kost, C.; Patz, C.-D.; Ristow, R.; Wachter, H. Quantitation of compounds in wine using 1H NMR spectroscopy: Description of the method and collaborative study. J. AOAC Int. 2016, 99, 1295–1304. [Google Scholar] [CrossRef]
- Son, H.-S.; Hwang, G.-S.; Kim, K.M.; Ahn, H.-J.; Park, W.-M.; Van Den Berg, F.; Hong, Y.-S.; Lee, C.-H. Metabolomic studies on geographical grapes and their wines using 1H NMR analysis coupled with multivariate statistics. J. Agric. Food. Chem. 2009, 57, 1481–1490. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Zhong, Q.; Fauhl-Hassek, C.; Pfister, M.K.H.; Horn, B.; Huang, Z. Classification of Chinese wine varieties using 1H NMR spectroscopy combined with multivariate statistical analysis. Food Control 2018, 88, 113–122. [Google Scholar] [CrossRef]
- Caruso, M.; Galgano, F.; Castiglione Morelli, M.A.; Viggiani, L.; Lencioni, L.; Giussani, B.; Favati, F. Chemical profile of white wines produced from ‘Greco bianco’ grape variety in different Italian areas by nuclear magnetic resonance (NMR) and conventional physicochemical analyses. J. Agric. Food. Chem. 2012, 60, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Pereira, G.E.; Gaudillère, J.P.; Van Leeuwen, C.; Hilbert, G.; Maucourt, M.; Deborde, C.; Moing, A.; Rolin, D. 1H-NMR metabolic profiling of wines from three cultivars, three soil types and two contrasting vintages. J. Int. Sci. Vigne Vin 2007, 41, 103–109. [Google Scholar]
- Lee, J.-E.; Hwang, G.-S.; Van Den Berg, F.; Lee, C.-H.; Hong, Y.-S. Evidence of vintage effects on grape wines using 1H NMR-based metabolomic study. Anal. Chim. Acta 2009, 648, 71–76. [Google Scholar] [CrossRef]
- Cassino, C.; Tsolakis, C.; Bonello, F.; Gianotti, V.; Osella, D. Wine evolution during bottle aging, studied by 1H NMR spectroscopy and multivariate statistical analysis. Food Res. Int. 2018, 116, 566–577. [Google Scholar] [CrossRef]
- Amargianitaki, M.; Spyros, A. NMR-based metabolomics in wine quality control and authentication. Chem. Biol. Technol. Agric. 2017, 4, 9. [Google Scholar] [CrossRef] [Green Version]
- Alves Filho, E.G.; Silva, L.M.A.; Ribeiro, P.R.V.; de Brito, E.S.; Zocolo, G.J.; Souza-Leão, P.C.; Marques, A.T.B.; Quintela, A.L.; Larsen, F.H.; Canuto, K.M. 1H NMR and LC-MS-based metabolomic approach for evaluation of the seasonality and viticultural practices in wines from São Francisco River Valley, a Brazilian semi-arid region. Food Chem. 2019, 289, 558–567. [Google Scholar] [CrossRef]
- López-Rituerto, E.; Savorani, F.; Avenoza, A.; Busto, J.H.; Peregrina, J.M.; Engelsen, S.B. Investigations of la Rioja terroir for wine production using 1H NMR metabolomics. J. Agric. Food. Chem. 2012, 60, 3452–3461. [Google Scholar] [CrossRef] [PubMed]
- Mazzei, P.; Spaccini, R.; Francesca, N.; Moschetti, G.; Piccolo, A. Metabolomic by 1H NMR spectroscopy differentiates “Fiano Di Avellino” white wines obtained with different yeast strains. J. Agric. Food. Chem. 2013, 61, 10816–10822. [Google Scholar] [CrossRef] [Green Version]
- De Pascali, S.A.; Coletta, A.; Del Coco, L.; Basile, T.; Gambacorta, G.; Fanizzi, F.P. Viticultural practice and winemaking effects on metabolic profile of Negroamaro. Food Chem. 2014, 161, 112–119. [Google Scholar] [CrossRef] [PubMed]
- Baiano, A.; Terracone, C.; Longobardi, F.; Ventrella, A.; Agostiano, A.; Del Nobile, M.A. Effects of different vinification technologies on physical and chemical characteristics of Sauvignon blanc wines. Food Chem. 2012, 135, 2694–2701. [Google Scholar] [CrossRef]
- González-Neves, G.; Favre, G.; Gil, G. Effect of fining on the colour and pigment composition of young red wines. Food Chem. 2014, 157, 385–392. [Google Scholar] [CrossRef]
- Doco, T.; Williams, P.; Cheynier, V. Effect of flash Release and pectinolytic enzyme treatments on wine polysaccharide composition. J. Agric. Food. Chem. 2007, 55, 6643–6649. [Google Scholar] [CrossRef]
- Parish, K.J.; Herbst-Johnstone, M.; Bouda, F.; Klaere, S.; Fedrizzi, B. Pre-fermentation fining effects on the aroma chemistry of Marlborough Sauvignon blanc press fractions. Food Chem. 2016, 208, 326–335. [Google Scholar] [CrossRef] [PubMed]
- Arnous, A.; Meyer, A.S. Discriminated release of phenolic substances from red wine grape skins (Vitis vinifera L.) by multicomponent enzymes treatment. Biochem. Eng. J. 2010, 49, 68–77. [Google Scholar] [CrossRef]
- Wheelock, Å.M.; Wheelock, C.E. Trials and tribulations of ‘omics data analysis: Assessing quality of SIMCA-based multivariate models using examples from pulmonary medicine. Mol. BioSyst. 2013, 9, 2589–2596. [Google Scholar] [CrossRef] [Green Version]
- Benbouguerra, N.; Valls-Fonayet, J.; Krisa, S.; Garcia, F.; Saucier, C.; Richard, T.; Hornedo-Ortega, R. Polyphenolic characterization of Merlot, Tannat and Syrah skin extracts at different degrees of maturity and anti-inflammatory potential in RAW 264.7 cells. Foods 2021, 10, 541. [Google Scholar] [CrossRef]
- Ferrero-del-Teso, S.; Arias, I.; Escudero, A.; Ferreira, V.; Fernández-Zurbano, P.; Sáenz-Navajas, M.-P. Effect of grape maturity on wine sensory and chemical features: The case of Moristel wines. LWT 2020, 118, 108848. [Google Scholar] [CrossRef]
- Bindon, K.; Varela, C.; Kennedy, J.; Holt, H.; Herderich, M. Relationships between harvest time and wine composition in Vitis vinifera L. cv. Cabernet Sauvignon 1. Grape and wine chemistry. Food Chem. 2013, 138, 1696–1705. [Google Scholar] [CrossRef]
- Lee, J.E.; Hong, Y.S.; Lee, C.H. Characterization of fermentative behaviors of lactic acid bacteria in grape wines through 1H NMR- and GC-based metabolic profiling. J. Agric. Food. Chem. 2009, 57, 4810–4817. [Google Scholar] [CrossRef]
- Long, D.; Wilkinson, K.L.; Poole, K.; Taylor, D.K.; Warren, T.; Astorga, A.M.; Jiranek, V. Rapid method for proline determination in grape juice and wine. J. Agric. Food. Chem. 2012, 60, 4259–4264. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, O.; Sepúlveda, M.; Schwartz, M. Effects of enzymatic treatment on anthocyanic pigments from grapes skin from chilean wine. Food Chem. 2004, 87, 487–490. [Google Scholar] [CrossRef]
- Li, S.; Bindon, K.; Bastian, S.E.P.; Jiranek, V.; Wilkinson, K.L. Use of winemaking supplements to modify the composition and sensory properties of Shiraz wine. J. Agric. Food. Chem. 2017, 65, 1353–1364. [Google Scholar] [CrossRef] [PubMed]
- Hufnagel, J.C.; Hofmann, T. Quantitative reconstruction of the nonvolatile sensometabolome of a red wine. J. Agric. Food. Chem. 2008, 56, 9190–9199. [Google Scholar] [CrossRef]
- Ducasse, M.-A.; Canal-Llauberes, R.-M.; de Lumley, M.; Williams, P.; Souquet, J.-M.; Fulcrand, H.; Doco, T.; Cheynier, V. Effect of macerating enzyme treatment on the polyphenol and polysaccharide composition of red wines. Food Chem. 2010, 118, 369–376. [Google Scholar] [CrossRef]
- Thoukis, G.; Ueda, M.; Wright, D. The formation of succinic acid during alcoholic fermentation. Am. J. Enol. Vitic. 1965, 16, 1–8. [Google Scholar]
- De-la-Fuente-Blanco, A.; Sáenz-Navajas, M.-P.; Ferreira, V. On the effects of higher alcohols on red wine aroma. Food Chem. 2016, 210, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Marangon, M.; Vincenzi, S.; Curioni, A. Wine Fining with Plant Proteins. Molecules 2019, 24, 2186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gil, M.; Louazil, P.; Iturmendi, N.; Moine, V.; Cheynier, V.; Saucier, C. Effect of polyvinylpolypyrrolidone treatment on rosés wines during fermentation: Impact on color, polyphenols and thiol aromas. Food Chem. 2019, 295, 493–498. [Google Scholar] [CrossRef]
- Maury, C.; Sarni-Manchado, P.; Cheynier, V. Highlighting protein fining residues in a model red wine. Food Chem. 2019, 279, 272–278. [Google Scholar] [CrossRef]
- Jackson, R.S. Chapter 6—Chemical constituents of grapes and wine. In Wine Science, 5th ed.; Jackson, R.S., Ed.; Academic Press: Cambridge, MA, USA, 2020; pp. 375–459. [Google Scholar]
- Ma, T.-Z.; Gong, P.-F.; Lu, R.-R.; Zhang, B.; Morata, A.; Han, S.-Y. Effect of different clarification treatments on the volatile composition and aromatic attributes of ‘Italian Riesling’ icewine. Molecules 2020, 25, 2657. [Google Scholar] [CrossRef]
- Rousserie, P.; Lacampagne, S.; Vanbrabant, S.; Rabot, A.; Geny-Denis, L. Wine tannins: Where are they coming from? A method to access the importance of berry part on wine tannins content. MethodsX 2020, 7, 100961. [Google Scholar] [CrossRef]
- Cobas, C.; Seoane, F.; Domínguez, S.; Sykora, S.; Davies, A.N. A new approach to improving automated analysis of proton NMR spectra through Global Spectral Deconvolution (GSD). Spectrosc. Eur. 2011, 23, 26–30. [Google Scholar]
- Tyagi, R.; Maan, K.; Khushu, S.; Rana, P. Urine metabolomics based prediction model approach for radiation exposure. Sci. Rep. 2020, 10, 16063. [Google Scholar] [CrossRef] [PubMed]
- Westerhuis, J.A.; van Velzen, E.J.J.; Hoefsloot, H.C.J.; Smilde, A.K. Multivariate paired data analysis: Multilevel PLSDA versus OPLSDA. Metabolomics 2010, 6, 119–128. [Google Scholar] [CrossRef] [Green Version]
- Trygg, J.; Wold, S. Orthogonal projections to latent structures (O-PLS). J. Chemom. 2002, 16, 119–128. [Google Scholar] [CrossRef]
- Ruiying, C.; Zeyun, L.; Yongliang, Y.; Zijia, Z.; Ji, Z.; Xin, T.; Xiaojian, Z. A comprehensive analysis of metabolomics and transcriptomics in non-small cell lung cancer. PLoS ONE 2020, 15, e0232272. [Google Scholar] [CrossRef]
- Gougeon, L.; da Costa, G.; Richard, T.; Guyon, F. Wine authenticity by quantitative 1 H NMR versus multitechnique analysis: A case study. Food Anal. Methods 2019, 12, 956–965. [Google Scholar] [CrossRef]




| Compound | δ1H (Multiplicity, J in Hz, Assignment) | |
|---|---|---|
| 1 | leucine | 0.96 (d, 6.2, 2CH3), 1.71 (m, CHCH2), 3.74 (m, CH) |
| 2 | isoleucine | 0.93 (t, 7.4, CH3), 0.99 (d, 7.0, CH3), 1.24 (m, CH2), 1.45 (m, CH2), 1.97 (m, CH), 3.66 (d, 3.9, CH) |
| 3 | valine | 0.99 (d, 7.3, CH3), 1.04 (d, 7.3, CH3), 2.28 (m, CH), 3.66 (d, 4.3, CH) |
| 4 | 2,3-butanediol | 1.13 (d, 6.2, 2CH3), 3.61 (m, 2CH) |
| 5 | ethanol | 1.17 (t, 7.2, CH3), 3.65 (q, CH2) |
| 6 | threonine | 1.32 (d, 6.7, CH3), 2.58 (d, 4.9, CH), 4.24 (m, CH) |
| 7 | acetoin | 1.37 (d, 7.0, CH3), 2.21 (s, CH3), 4.42 (q, CH) |
| 8 | lactic acid | 1.40 (d, 7.0, CH3), 4.31 (q, 7.0, CH) |
| 9 | alanine | 1.50 (d, 7.2, CH3), 3.76 (q, CH) |
| 10 | isopentanol | 0.88 (d, 6.7, 2CH3), 1.44 (q, CH2); 1.66 (m, CH), 3.61 (t, 6.7, CH2) |
| 11 | arginine | 1.70 (m, CH2), 1.89 (m, CH2), 3.23 (t, CH2), 3.75 (t, 6.5, CH) |
| 12 | proline | 1.99 (m, CH2), 2.06 (m, CH), 2.33 (m, CH), 3.32 (dt, 14.0, 7.1, CH), 3.42 (dt, 11.6 and 7.0, CH), 4.11 (dd, 8.6 and 6.4, CH) |
| 13 | ethyl acetate | 1.26 (t, 7.2, CH3), 4.12 (q, CH2), 2.07 (s, CH3) |
| 14 | acetic acid | 2.08 (s, CH3) |
| 15 | ethanal | 2.23 (d, 3.0, CH3), 9.67 (q, CH) |
| 16 | pyruvic acid | 2.35 (s, CH3) |
| 17 | γ-aminobutyric acid | 1.96 (m, CH2), 2.50 (t, 7.3, CH2), 3.05 (m, CH2) |
| 18 | succinic acid | 2.65 (s, 2CH2) |
| 19 | malic acid | 2.78 (dd, 16.3 and 7.0, CH), 2.89 (dd, 16.3 and 4.5, CH), 4.53 (dd, CH) |
| 20 | citric acid | 2.79 (d, 15.6, CH2), 2.94 (d, 15.6, CH2) |
| 21 | choline | 3.19 (s, 3CH3), 3.51 (dd, CH2), 4.05 (m, CH2) |
| 22 | myo-inositol | 3.27 (t, 9.7, CH), 3.52 (dd, 10.0 and 2.8, 2CH), 3.61 (t, 2.8, 2CH), 4.05 (t, 2.8, CH) |
| 23 | methanol | 3.35 (s, CH3) |
| 24 | isobutanol | 0.87 (d, 6.7, 2CH3), 1,73 (m, CH), 3.36 (d, 6.7, CH2) |
| 25 | glycerol | 3.55 (dd, 11.8 and 6.5, CH2), 3.64 (dd, CH2), 3.77 (m, CH) |
| 26 | mannitol | 3.65 (dd, 11.7, 6.2 CH2), 3.73 (m, CH), 3.77 (d, 9.0, CH), 3.84 (dd, 11.9, 2.8, CH2) |
| 27 | fructose | 3.56 (m, CH2), 3.70 (m, 2CH2), 3.77 (m, CHCHCH2), 3.87 (dd, 9.9, 3.4, CH), 3.97 (m, CH), 4.00 (dd, 12.8, 1.0 CH2), 4.09 (m, 2CH) |
| 28 | ethyl lactate | 1.28 (t, CH3), 1.42 (d, 7.0, CH3), 4.22 (q, 7.06, CH), 4.39 (q, 7.0, CH) |
| 29 | arabinose | 3.51 (dd, CH), 3.68 (m, CHCH2), 3.83 (dd, CH), 3.90 (m, CHCH2), 3.95(m, CH), 4.02 (m, CHCH2), 4.50 (d, 7.7, CH), 5.25 (d, CH) |
| 30 | glucose | 3.23 (dd, 9.2, 8.0, CH), 3.39 (m, CH), 3.45 (dd, 9.8, 3.7, CH), 3.72 (m, CHCH2), 3.82 (m, CHCH2), 3.88 (dd, 12.2, 2.1, CH2), 4.63 (d, 7.9, CH), 5.22 (d, 3.6, CH) |
| 31 | tartaric acid | 4.60 (s, 2CH) |
| 32 | xylose | 3.21 (dd, 9.3, 7.9, CH), 3.31 (t, 11.4, CH2), 3.42 (t, 9.25, CH), 3.51 (dd, 9.3, 3.7, CH), 3.63 (m, CHCHCH2), 3.91 (dd, 11.5, 5.5, CH2), 4.57 (d, 7.9, CH), 5.19 (d, 3.7, CH) |
| 33 | galacturonic acid | 3.49 (dd, 8.0, 10.0, CH), 3.69 (dd, 9, 3.5, CH), 3.80 (dd, 10.3, 3.8, CH), 3,92 (dd, 10.3, 3.4, CH), 4.24 (dd, 3.6, 1.2, CH), 4.26 (d, 1.2, CH), 4.31 (dd, 3.3, 1.4, CH), 5.32 (d, 3.8, CH) |
| 34 | glucuronic acid | 3.29 (t, 8.6, CH), 3.51 (m, 2CH), 3.58 (dd, 9.7, 3.7, CH), 3.73 (m, 2CH), 4.08 (d, 10.8, CH), 4.64 (d, 7.9, CH), 5.25 (d, 3.7, CH), 5.55 (d, 4, CH) |
| 35 | sorbic acid | 1.82 (d, 6.2, CH3), 5.78 (d, 15.3, CH), 6.25 (m, 2CH), 7.16 (dd, 15.3, 10.3, CH) |
| 36 | epicatechin | 2.76 (m, CH2), 2.90 (m, CH2), 4.32 (m, CH), 4.95 (m, CH), 6.09 (d, 2.0, CH), 6.12 (d, 2.0, CH), 6.93 (m, CH2), 7.03 (d, 2.0, CH) |
| 37 | catechin | 2.53 (dd, CH2), 2.85 (m, CH2), 4.15 (m, CH), 4.41 (d, 7.0, CH), 5.99 (d, 2.0, CH), 6.08 (d, 2.3, CH), 6.84 (d, 8.6, CH), 6.92 (m, 2CH) |
| 38 | caffeic acid | 6.33 (d, 16.0, CH), 6.92 (d, 8.0, CH), 7.07 (dd, 8.2, 2.0, CH), 7.14 (d, 2.0, CH), 7.29 (d, CH) |
| 39 | fumaric acid | 6.78 (s, 2CH) |
| 40 | shikimic acid | 2.21 (dd, 18.2,7.0, CH2), 2.75 (dd, 18.0, 5.3, CH2), 3.74 (dd, 8.6, 4.3, CH), 4.01 (m, CH), 4.42 (t, 4.1, CH), 6.82 (dt, CH) |
| 41 | tyrosol | 2.77 (t, CH2), 3.77 (t, CH2), 6.84 (m, 8.4, 2CH), 7.17 (m, 8.4, 2CH) |
| 42 | tyrosine | 3.02 (dd, CH2), 3.17 (dd, CH2), 3.92 (dd, CH), 6.86 (m, 8.4, 2CH), 7.17 (m, 8.6, 2CH) |
| 43 | gallic acid | 7.16 (s, 2CH) |
| 44 | phenethyl alcohol | 2.85 (t, 6.62, CH2), 3.74 (t, CH2), 7.33 (m, 5CH) |
| 45 | syringic acid | 3.84 (s, 2CH3), 7.36 (s, 2CH) |
| 46 | histidine | 3.16 (dd, 15.6, 7.7, CH), 3.23 (dd, 16.0, 5.0, CH), 3.98 (dd, 7.7, 5.0, CH), 7.09 (d, 5.0, CH), 7.90 (d, 1.1, CH) |
| 47 | trigonelline | 4.42 (s, CH3), 8.07 (m, CH), 8.82 (m, 2CH), 9.11 (s, CH) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Le Mao, I.; Martin-Pernier, J.; Bautista, C.; Lacampagne, S.; Richard, T.; Da Costa, G. 1H-NMR Metabolomics as a Tool for Winemaking Monitoring. Molecules 2021, 26, 6771. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules26226771
Le Mao I, Martin-Pernier J, Bautista C, Lacampagne S, Richard T, Da Costa G. 1H-NMR Metabolomics as a Tool for Winemaking Monitoring. Molecules. 2021; 26(22):6771. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules26226771
Chicago/Turabian StyleLe Mao, Inès, Jean Martin-Pernier, Charlyne Bautista, Soizic Lacampagne, Tristan Richard, and Gregory Da Costa. 2021. "1H-NMR Metabolomics as a Tool for Winemaking Monitoring" Molecules 26, no. 22: 6771. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules26226771