A DFT/PCM Study on the Affinity of Salinomycin to Bind Monovalent Metal Cations
Abstract
:1. Introduction
2. Results and Discussion
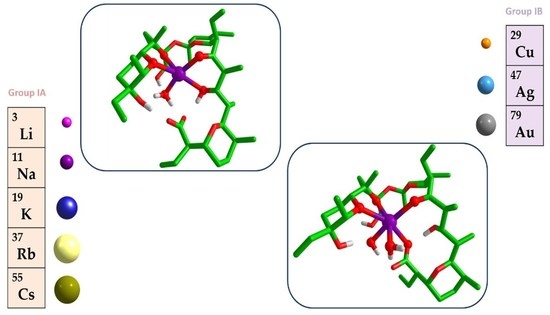
2.1. Salinomycin Complexes with Alkali Metal Ions
2.2. Salinomycin Complexes with Coinage Metal Ions
2.3. Salinomycin vs. Monensin
3. Methods
3.1. Gibbs Free Energy Change for the Reaction Modeled
− ΔGεsolv ([SalNa]) − ΔGεsolv ([M+-solution]).
3.2. DFT/PCM Calculations
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Miyazaki, Y.; Shibuya, M.; Sugawara, H.; Kawaguchi, O.; Hirsoe, C. Salinomycin, a new polyether antibiotic. J. Antibiot. 1974, 27, 814–821. [Google Scholar] [CrossRef]
- Chappel, L.R. The site of action of the anticoccidial salinomycin (Coxistac). J. Parasitol. 1979, 65, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Long, P.L.; Jeffers, T.K. Studies on the stage of action of ionophorous antibiotics against Eimeria. J. Parasitol. 1982, 68, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Coombs, G.H.; Müller, S. Recent advances in the search for new anti-coccidial drugs. Int. J. Parasitol. 2002, 32, 497–508. [Google Scholar] [CrossRef]
- Mahmoudi, N.; de Julian-Ortiz, J.-V.; Ciceron, L.; Galvez, J.; Mazier, D.; Danis, M.; Derouin, F.; Garcıa-Domenech, R. Identification of new antimalarial drugs by linear discriminant analysis and topological virtual screening. J. Antimicrob. Chemother. 2006, 57, 489–497. [Google Scholar] [CrossRef] [PubMed]
- D’Alessandro, S.; Corbett, Y.; Ilboudo, D.P.; Misiano, P.; Dahiya, N.; Abay, S.M.; Habluetzel, A.; Grande, R.; Gismondo, M.R.; Dechering, K.J.; et al. Salinomycin and other ionophores as a new class of antimalarial drugs with transmission-blocking activity. Antimicrob. Agents Chemother. 2015, 59, 5135–5144. [Google Scholar] [CrossRef] [Green Version]
- Yuan, C.; Huang, X.; Zhai, R.; Ma, Y.; Xu, A.; Zhang, P.; Yang, Q. In Vitro antiviral activities of salinomycin on porcine epidemic diarrhea virus. Viruses 2021, 13, 580. [Google Scholar] [CrossRef]
- Gupta, P.B.; Onder, T.T.; Jiang, G.; Tao, K.; Kuperwasser, C.; Weinberg, R.A.; Lander, E.S. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell 2009, 138, 645–659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, D.; Choi, M.Y.; Yu, J.; Castro, J.E.; Kipps, T.J.; Carson, D.A. Salinomycin inhibits Wnt signaling and selectively induces apoptosis in chronic lymphocytic leukemia cells. Proc. Natl. Acad. Sci. USA 2011, 108, 13253–13257. [Google Scholar] [CrossRef] [Green Version]
- Tang, Q.L.; Zhao, Z.Q.; Li, J.C.; Liang, Y.; Yin, J.Q.; Zou, C.Y.; Xie, X.B.; Zeng, Y.X.; Shen, J.N.; Kang, T.; et al. Salinomycin inhibits osteosarcoma by targeting its tumor stem cells. Cancer Lett. 2011, 311, 113–121. [Google Scholar] [CrossRef]
- Naujokat, C.; Steinhart, R. Salinomycin as a drug for targeting human cancer stem cells. J. Biomed. Biotechnol. 2012, 2012, 950658. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Wang, F.; Wong, E.T.; Fonkem, E.; Hsieh, T.C.; Wu, J.M.; Wu, E. Salinomycin: A novel anti-cancer agent with known anti-coccidial activities. Curr. Med. Chem. 2013, 20, 4095–4101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.; Tian, Y.; Song, F.; Fu, C.; Han, B.; Wang, Y. Salinomycin inhibits the growth of colorectal carcinoma by targeting tumor stem cells. Oncol. Rep. 2015, 34, 2469–2476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Najumudeen, A.K.; Jaiswal, A.; Lectez, B.; Oetken-Lindholm, C.; Guzman, C.; Siljamaki, E.; Posada, I.M.D.; Lacey, E.; Aittkallio, T.; Abankwa, D. Cancer stem cell drugs target K-ras signaling in a stemness context. Oncogene 2016, 35, 5248–5262. [Google Scholar] [CrossRef] [Green Version]
- Dewangan, J.; Srivastava, S.; Rath, S.K. Salinomycin: A new paradigm in cancer therapy. Tumour. Biol. 2017, 39, 1010428317695035. [Google Scholar] [CrossRef] [Green Version]
- Mai, T.T.; Hamai, A.; Hienzsch, A.; Caneque, T.; Muller, S.; Wicinski, J.; Cabaud, O.; Leroy, C.; David, A.; Acevedo, V.; et al. Salinomycin kills cancer stem cells by sequestering iron in lysosomes. Nat. Chem. 2017, 9, 1025–1033. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.G.; Shin, S.J.; Chung, H.W.; Kwon, S.H.; Cha, S.D.; Lee, J.E.; Cho, C.H. Salinomycin reduces stemness and induces apoptosis on human ovarian cancer stem cell. J. Gynecol. Oncol. 2017, 28, e14. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Wang, Q.; Mao, J.; Qin, T.; Sun, Y.; Yang, J.; Han, Y.; Li, L.; Li, Q. Salinomycin suppresses cancer cell stemness and attenuates TGF-beta-induced epithelial-mesenchymal transition of renal cell carcinoma cells. Chem. Biol. Interact. 2018, 296, 145–153. [Google Scholar] [CrossRef]
- Zhou, J.; Liu, S.; Wang, Y.; Dai, W.; Zou, H.; Wang, S.; Zhang, J.; Pan, J. Salinomycin effectively eliminates cancer stem-like cells and obviates hepatic metastasis in uveal melanoma. Mol. Cancer 2019, 18, 159. [Google Scholar] [CrossRef]
- Liu, Y.; Hao, Y.; Li, Y.; Zheng, Y.; Dai, J.; Zhong, F.; Wei, W.; Fang, Z. Salinomycin induces autophagic cell death in salinomycin-sensitive melanoma cells through inhibition of autophagic flux. Sci. Rep. 2020, 10, 18515. [Google Scholar] [CrossRef]
- Li, K.; Pang, L.; Pan, X.; Fan, S.; Wang, X.; Wang, Q.; Dai, P.; Gao, W.; Gao, J. GE11 modified PLGA/TPGS nanoparticles targeting delivery of salinomycin to breast cancer cells. Technol. Cancer Res. Treat. 2021, 20, 15330338211004954. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhang, H.; Zhu, Y.; Wu, Z.; Cui, C.; Cai, F. Anticancer mechanisms of salinomycin in breast cancer and its clinical applications. Front. Oncol. 2021, 11, 654428. [Google Scholar] [CrossRef] [PubMed]
- Pinkerton, M.; Stenrauf, L.K. Molecular structure of monovalent metal cation complexes of monensin. J. Mol. Biol. 1970, 49, 533–546. [Google Scholar] [CrossRef]
- Lutz, W.K.; Winkler, F.K.; Dunitz, J.D. Crystal structure of the antibiotic monensin. Similarities and differences between free acid and metal complex. Helv. Chim. Acta 1971, 54, 1103–1108. [Google Scholar] [CrossRef]
- Duax, W.L.; Smith, G.D.; Strong, P.D. Complexation of metal ions by monensin. Crystal and molecular structure of hydrated and anhydrous crystal forms of sodium monensin. J. Am. Chem. Soc. 1980, 102, 6725–6729. [Google Scholar] [CrossRef]
- Walba, D.M.; Hermsmeier, M.; Haltiwanger, R.C.; Noordik, J.H. Crystal structures of monensin B lithium and silver salts. J. Org. Chem. 1986, 51, 245–247. [Google Scholar] [CrossRef]
- Pangborn, W.; Duax, W.; Langs, D. The hydrated potassium complex of the ionophore monensin A. J. Am. Chem. Soc. 1987, 109, 2163–2165. [Google Scholar] [CrossRef]
- Huczynski, A.; Ratajczak-Sitarz, M.; Katrusiak, A.; Brzezinski, B. Molecular structure of the 1:1 inclusion complex of monensin A sodium salt with acetonitrile. J. Mol. Struct. 2007, 832, 84–89. [Google Scholar] [CrossRef]
- Huczynski, A.; Ratajczak-Sitarz, M.; Katrusiak, A.; Brzezinski, B. Molecular structure of the 1:1 inclusion complex of monensin A lithium salt with acetonitrile. J. Mol. Struct. 2007, 871, 92–97. [Google Scholar] [CrossRef]
- Huczynski, A.; Ratajczak-Sitarz, M.; Katrusiak, A.; Brzezinski, B. Molecular structure of rubidium six-coordinated dihydrate complex with monensin A. J. Mol. Struct. 2008, 883, 224–229. [Google Scholar] [CrossRef]
- Paulus, E.F.; Kurz, M.; Matter, H.; Vertesy, L. Solid-state and solution structure of the salinomycin-sodium complex: Stabilization of different conformers for an ionophore in different environments. J. Am. Chem. Soc. 1998, 120, 8209–8221. [Google Scholar] [CrossRef]
- Mitani, M.; Yamanishi, T.; Miyazaki, Y. Salinomycin: A new monovalent cation ionophore. Biochem. Biophys Res. Comm. 1975, 66, 12311236. [Google Scholar] [CrossRef]
- Shannon, R.D. Revised effective ionic radii and systematic studies in interatomic distances in halides and chalcogenides. Acta Cryst. 1976, A32, 751–767. [Google Scholar] [CrossRef]
- Gagne, O.C.; Hawthorne, F.C. Empirical Lewis acid strengths for 135 cations bonded to oxygen. Acta Cryst. B 2017, 73, 956–961. [Google Scholar] [CrossRef] [PubMed]
- Dudev, T.; Cheshmedzhieva, D.; Dimitrova, R.; Dorkov, P.; Pantcheva, I. Factors governing the competition between group IA and IB cations for monensin A: A DFT/PCM study. RSC Adv. 2020, 10, 5734–5741. [Google Scholar] [CrossRef]
- Laguna, A. Modern Supramolecular Gold Chemistry: Gold-Metal Interactions and Applications, 1st ed.; WILEY-VCH Verlag GmbH & Co. KgaA: Weinheim, Germany, 2008. [Google Scholar]
- Angelova, S.E.; Nikolova, V.K.; Dudev, T.M. Determinants of the host-guest interactions between α-, β- and γ-cyclodextrins and group IA, IIA and IIIA metal cations: A DFT/PCM study. Phys. Chem. Chem. Phys. 2017, 19, 15129–15136. [Google Scholar] [CrossRef]
- Dudev, T.; Lim, C. Determinants of K+ vs. Na+ selectivity in potassium channels. J. Am. Chem. Soc. 2009, 131, 8092–8101. [Google Scholar] [CrossRef] [PubMed]
- Dudev, T.; Lim, C. Ion selectivity strategies of sodium channel selectivity filters. Acc. Chem. Res. 2014, 47, 3580–3587. [Google Scholar] [CrossRef] [PubMed]
- Dudev, T.; Cheshmedzhieva, D.; Doudeva, L. Competition between abiogenic Al3+ and native Mg2+, Fe2+ and Zn2+ ions in protein binding sites: Implications for aluminum toxicity. J. Mol. Model. 2018, 24, 55. [Google Scholar] [CrossRef]
- Dudev, T.; Grauffel, C.; Lim, C. How Pb2+ binds and modulates properties of Ca2+-signaling proteins. Inorg. Chem. 2018, 57, 14798–14809. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Montgomery, J.A., Jr.; Vreven, T.; Kudin, K.N.; Burant, J.C.; et al. Gaussian 09; Gaussian Inc.: Pittsburgh, PA, USA, 2009. [Google Scholar]
- Marenich, A.V.; Cramer, C.J.; Truhlar, D.G. Universal solvation model based on solute electron density and on a continuum model of the solvent defined by the bulk dielectric constant and atomic surface tensions. J. Phys. Chem. B 2009, 113, 63786396. [Google Scholar] [CrossRef] [PubMed]
- Kircheva, N.; Dudev, T. Novel insights into gallium’s mechanism of therapeutic action: A DFT/PCM study of the interaction between Ga3+ and ribonucleotide reductase substrates. J. Phys. Chem. B 2019, 123, 5444–5451. [Google Scholar] [CrossRef] [PubMed]





| Cation | Ionic Radius (r, Å) [33] 1 | Charge Density 2 (e/Å3) | Lewis Acidity (Valence Units) [34] | Charge Transfer to the Metal 3 (e) |
|---|---|---|---|---|
| Li+ | 0.59 (IV) | 1.16 | 0.215 | 0.286/0.260 |
| Na+ | 1.00/1.02 (V/VI) | 0.24/0.22 | 0.159 | 0.215/0.198 |
| K+ | 1.38 (VI) | 0.09 | 0.108 | 0.166/0.152 |
| Rb+ | 1.52/1.56 (VI/VII) | 0.07/0.06 | 0.099 | 0.142/0.132 |
| Cs+ | -/1.74 (VII/VIII) | -/0.04 | 0.084 | 0.129/0.119 |
| Cu+ | 0.60 (IV) | 1.10 | 0.400 | 0.360/0.338 |
| Ag+ | 1.09/1.15 (V/VI) | 0.18/0.16 | 0.191 | 0.300/0.281 |
| Au+ | -/1.37 (II/VI) | -/0.09 | - | 0.412/0.381 |
| Reaction | ΔGε | Reaction | ΔGε |
|---|---|---|---|
| [Li+-solution] + [SalNa-W1] → [SalLi-W1] + [Na+-solution] | ΔG1 = −21.6 ΔG2 = −7.8 ΔG4 = −3.2 ΔG32 = 1.5 ΔG78 = 1.4 | [Li+-solution] + [SalNa-W2] → [SalLi-W2] + [Na+-solution] | ΔG1 = −24.0 ΔG2 = −10.6 ΔG4 = −6.5 ΔG32 = −1.9 ΔG78 = −1.7 |
| [K+-solution] + [SalNa-W1] → [SalK-W1] + [Na+-solution] | ΔG1 = 19.8 ΔG2 = 9.9 ΔG4 = 6.8 ΔG32 = 2.8 ΔG78 = 2.8 | [K+-solution] + [SalNa-W2] → [SalK-W2] + [Na+-solution] | ΔG1 = 20.6 ΔG2 = 10.0 ΔG4 = 6.3 ΔG32 = 2.7 ΔG78 = 2.7 |
| [Rb+-solution] + [SalNa-W1] → [SalRb-W1] + [Na+-solution] | ΔG1 = 58.2 ΔG2 = 46.2 ΔG4 = 41.6 ΔG32 = 34.3 ΔG78 = 33.2 | [Rb+-solution] + [SalNa-W2] → [SalRb-W2] + [Na+-solution] | ΔG1 = 56.1 ΔG2 = 43.5 ΔG4 = 38.2 ΔG32 = 32.8 ΔG78 = 31.2 |
| [Cs+-solution] + [SalNa-W1] → [SalCs-W1] + [Na+-solution] | ΔG1 = 71.2 ΔG2 = 55.7 ΔG4 = 49.7 ΔG32 = 41.4 ΔG78 = 40.4 | [Cs+-solution] + [SalNa-W2] → [SalCs-W2] + [Na+-solution] | ΔG1 = 67.3 ΔG2 = 51.1 ΔG4 = 44.7 ΔG32 = 38.4 ΔG78 = 36.7 |
| [Cu+-solution] + [SalNa-W1] → [SalCu-W1] + [Na+-solution] | ΔG1 = −34.2 ΔG2 = −5.8 ΔG4 = 4.3 ΔG32 = 9.2 ΔG78 = 18.0 | [Cu+-solution] + [SalNa-W2] → [SalCu-W2] + [Na+-solution] | ΔG1 = −31.6 ΔG2 = −2.9 ΔG4 = 7.2 ΔG32 = 12.0 ΔG78 = 11.4 |
| [Ag+-solution] + [SalNa-W1] → [SalAg-W1] + [Na+-solution] | ΔG1 = 15.8 ΔG2 = 25.0 ΔG4 = 28.2 ΔG32 = 29.9 ΔG78 = 29.6 | [Ag+-solution] + [SalNa-W2] → [SalAg-W2] + [Na+-solution] | ΔG1 = 16.0 ΔG2 = 25.3 ΔG4 = 28.4 ΔG32 = 30.1 ΔG78 = 29.5 |
| [Au+-solution] + [SalNa-W1] → [SalAu-W1] + [Na+-solution] | ΔG1 = 5.2 ΔG2 = 20.0 ΔG4 = 24.8 ΔG32 = 27.5 ΔG78 = 26.8 | [Au+-solution] + [SalNa-W2] → [SalAu-W2] + [Na+-solution] | ΔG1 = 0.7 ΔG2 = 14.5 ΔG4 = 18.5 ΔG32 = 20.1 ΔG78 = 18.0 |
| Ion | qhirsh | qCM5 | qNBO |
|---|---|---|---|
| Li+ | 0.1746 | 0.5007 | 0.8892 |
| Na+ | 0.2600 | 0.6675 | 0.8993 |
| K+ | 0.2773 | 0.7816 | 0.9112 |
| Rb+ | 0.3023 | 0.8319 | 0.9284 |
| Cs+ | 0.3293 | 0.9555 | 0.9454 |
| Cu+ | 0.2780 | 0.4605 | 0.7379 |
| Ag+ | 0.3224 | 0.4651 | 0.6711 |
| Au+ | 0.2805 | 0.4202 | 0.5418 |
| Ion | qhirsh | qCM5 | qNBO |
|---|---|---|---|
| Li+ | 0.1725 | 0.5074 | 0.9005 |
| Na+ | 0.2474 | 0.6128 | 0.9121 |
| K+ | 0.2797 | 0.7392 | 0.9277 |
| Rb+ | 0.3263 | 0.7472 | 0.9395 |
| Cs+ | 0.3546 | 0.8293 | 0.9519 |
| Cu+ | 0.2411 | 0.4046 | 0.6690 |
| Ag+ | 0.3182 | 0.4450 | 0.6687 |
| Au+ | 0.2562 | 0.3946 | 0.4588 |
| SalM-W1 | ΔGsolv | |||
|---|---|---|---|---|
| Cyclohexane | Diethylether | Methanol | Water | |
| Li | −19.97 | −27.48 | −39.73 | −24.86 |
| Na | −19.10 | −26.06 | −37.70 | −22.72 |
| K | −18.49 | −25.17 | −36.98 | −22.24 |
| Rb | −19.29 | −26.42 | −40.78 | −26.00 |
| Cs | −19.42 | −26.75 | −40.97 | −26.20 |
| Cu | −19.73 | −27.12 | −44.50 | −20.88 |
| Ag | −19.49 | −26.74 | −39.99 | −25.21 |
| Au | −19.82 | −27.36 | −42.13 | −27.63 |
| SalM-W2 | ΔGsolv | |||
|---|---|---|---|---|
| Cyclohexane | Diethylether | Methanol | Water | |
| Li | −20.50 | −28.79 | −45.65 | −30.76 |
| Na | −19.23 | −26.52 | −43.59 | −27.90 |
| K | −19.31 | −26.91 | −43.83 | −28.26 |
| Rb | −20.05 | −28.07 | −46.11 | −31.25 |
| Cs | −20.17 | −28.27 | −45.98 | −31.21 |
| Cu | −19.53 | −27.17 | −50.19 | −35.30 |
| Ag | −19.51 | −27.09 | −45.93 | −30.65 |
| Au | −20.50 | −29.01 | −48.56 | −34.44 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dudev, T.; Cheshmedzhieva, D.; Dorkov, P.; Pantcheva, I. A DFT/PCM Study on the Affinity of Salinomycin to Bind Monovalent Metal Cations. Molecules 2022, 27, 532. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules27020532
Dudev T, Cheshmedzhieva D, Dorkov P, Pantcheva I. A DFT/PCM Study on the Affinity of Salinomycin to Bind Monovalent Metal Cations. Molecules. 2022; 27(2):532. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules27020532
Chicago/Turabian StyleDudev, Todor, Diana Cheshmedzhieva, Peter Dorkov, and Ivayla Pantcheva. 2022. "A DFT/PCM Study on the Affinity of Salinomycin to Bind Monovalent Metal Cations" Molecules 27, no. 2: 532. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules27020532