Probing Peptide and Protein Insertion in a Biomimetic S-Layer Supported Lipid Membrane Platform
Abstract
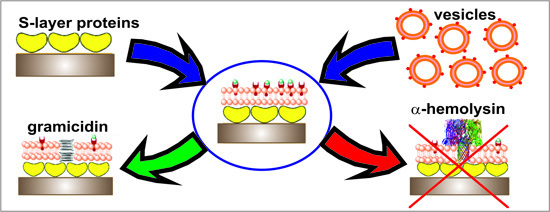
:1. Introduction

2. Results and Discussion
2.1. Assembly of S-Layer Supported Lipid Membranes

2.2. Electrical Characterization of S-Layer Supported Lipid Membranes
2.3. Incorporation of Gramicidin
2.4. Reconstitution of α-Hemolysin
3. Experimental Section
3.1. Isolation of S-Layer Proteins
3.2. EDC Activation
3.3. Vesicle Preparation
3.5. Quartz Crystal Microbalance with Dissipation Measurement (QCM-D)
3.6. Electrochemical Impedance Spectroscopy (EIS)
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Majed, S.; Yusko, E.C.; Billeh, Y.N.; Macrae, M.X.; Yang, J.; Mayer, M. Applications of biological pores in nanomedicine, sensing, and nanoelectronic. Curr. Opin. Biotechnol. 2010, 21, 439–476. [Google Scholar] [CrossRef]
- Hille, B. Ion Channels of Excitable Membranes, 3rd ed.; Sinauer Associates Inc.: Sunderland, MA, USA, 2001. [Google Scholar]
- Kaczorowski, G.; McManus, O.; Priest, B.; Garcia, M. Ion channels as drug targets: The next GPCRs. J. Gen. Physiol. 2008, 131, 399–405. [Google Scholar] [CrossRef]
- Früh, V.; Ijzerman, A.; Siegal, G. How to catch a membrane protein in action: A review of functional membrane protein immobilization strategies and their applications. Chem. Rev. 2011, 111, 640–656. [Google Scholar] [CrossRef] [PubMed]
- Schuster, B.; Sleytr, U.B. Biomimetic interfaces based on S-layer proteins, lipid membranes and functional biomolecules. J. R. Soc. Interface 2014. [Google Scholar] [CrossRef]
- Sackmann, E.; Tanaka, M. Supported membranes on soft polymer cushions: Fabrication, characterization and application. Trends Biotechnol. 2000, 18, 58–64. [Google Scholar] [CrossRef] [PubMed]
- Knoll, W.; Naumann, R.; Friedrich, M.; Robertson, J.W.F.; Lösche, M.; Heinrich, F.; McGillivray, D.J.; Schuster, B.; Gufler, P.C.; Pum, D.; et al. Solid supported lipid membranes: New concepts for the biomimetic functionalization of solid surfaces. Biointerphases 2008, 3, FA125–FA135. [Google Scholar] [CrossRef] [PubMed]
- Belegrinou, S.; Menon, S.; Dobrunz, D.; Meier, W. Solid-supported polymeric membranes. Soft Matter 2011, 7, 2202–2210. [Google Scholar] [CrossRef]
- Shen, H.; Lithgow, T.; Martin, L.L. Reconstitution of membrane proteins into model membranes: Seeking better ways to retain protein activities. Int. J. Mol. Sci. 2013, 14, 1589–1607. [Google Scholar] [CrossRef] [PubMed]
- Cooper, M. Advances in membrane receptor screening and analysis. J. Mol. Recognit. 2004, 17, 286–315. [Google Scholar] [CrossRef] [PubMed]
- Sleytr, U.B.; Schuster, B.; Egelseer, E.; Pum, D. S-layers: Principles and applications. FEMS Microbiol. Rev. 2014, 38, 823–864. [Google Scholar] [CrossRef] [PubMed]
- Sleytr, U.B. Heterologous reattachment of regular arrays of glycoproteins on bacterial surfaces. Nature 1975, 257, 400–402. [Google Scholar] [CrossRef] [PubMed]
- Sleytr, U.B.; Pum, D.; Egelseer, E.; Ilk, N.; Schuster, B. S-Layer Proteins. In Handbook of Biofunctional Surface; Knoll, W., Ed.; Pan Stanford Publishing: Singapore, 2013; pp. 507–568. [Google Scholar]
- Schuster, B.; Györvary, E.; Pum, D.; Sleytr, U.B. Nanotechnology with S-Layer Proteins. In Protein Nanotechnology: Protocols, Instrumentation and Applications; Book Series: Methods in Molecular Biology, 300; Vo-Dinh, T., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 101–124. [Google Scholar]
- Schuster, B.; Pum, D.; Sára, M.; Sleytr, U.B. S-layer proteins as key components of a versatile molecular construction kit for biomedical nanotechnology. Mini Rev. Med. Chem. 2006, 6, 909–920. [Google Scholar] [CrossRef] [PubMed]
- Sleytr, U.B.; Schuster, B.; Egelseer, E.M.; Pum, D.; Horejs, C.M.; Tscheliessnig, R.; Ilk, N. Nanotechnology with S-layer proteins as building blocks. Prog. Mol. Biol. Transl. Sci. 2010, 103, 277–352. [Google Scholar]
- Schuster, B.; Sleytr, U.B. Composite S-layer lipid structures. J. Struct. Biol. 2009, 168, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Schuster, B.; Sleytr, U.B. Nanotechnology with S-layer proteins. In Protein Nanotechnology; Gerrard, J.A., Ed.; Humana Press: Totowa, NJ, USA, 2013; pp. 153–157. [Google Scholar]
- Schuster, B.; Pum, D.; Sleytr, U.B. S-layer stabilized lipid membranes. Biointerphases 2008, 3, FA3–FA11. [Google Scholar] [CrossRef] [PubMed]
- Sleytr, U.B.; Schuster, B.; Pum, D. Nanotechnology and biomimetics with 2-D protein crystals. IEEE Eng. Med. Biol. 2003, 22, 140–150. [Google Scholar] [CrossRef]
- Schuster, B.; Pum, D.; Sara, M.; Braha, O.; Bayley, H.; Sleytr, U.B. S-layer ultrafiltration membranes: A new support for stabilizing functionalized lipid membranes. Langmuir 2001, 17, 499–503. [Google Scholar] [CrossRef]
- Schuster, B.; Weigert, S.; Pum, D.; Sara, M.; Sleytr, U.B. New method for generating tetraether lipid membranes on porous supports. Langmuir 2003, 19, 2392–2397. [Google Scholar] [CrossRef]
- Schuster, B.; Gufler, P.; Pum, D.; Sleytr, U.B. S-layer proteins as supporting scaffoldings for functional lipid membranes. IEEE Trans. Nanobiosci. 2004, 3, 16–21. [Google Scholar] [CrossRef]
- Albers, S.; Meyer, B. The archaeal cell envelope. Nat. Rev. Microbiol. 2011, 9, 414–426. [Google Scholar] [CrossRef]
- Andersson, M.; Jackman, J.; Wilson, D.; Jarvoll, P.; Alfredsson, V.; Okeyo, G.; Duran, R. Vesicle and bilayer formation of DPhPC and DPhPE mixtures and their bilayer’s electrical stability. Colloids Surf. B Biointerfaces 2011, 82, 550–561. [Google Scholar] [CrossRef] [PubMed]
- Hirn, R.; Schuster, B.; Sleytr, U.B.; Bayerl, T.M. The effect of S-layer protein adsorption and crystallization on the collective motion of a planar lipid bilayer studied by dynamic light scattering. Biophys. J. 1999, 77, 2066–2074. [Google Scholar] [CrossRef] [PubMed]
- Rossi, C.; Chopineau, J. Biomimtic tethered lipid membranes designed for membrane–protein interaction studies. Eur. Biophys. J. 2007, 36, 955–965. [Google Scholar] [CrossRef] [PubMed]
- Schrems, A.; Larisch, V.D.; Stanetty, C.; Dutter, K.; Damiati, S.; Sleytr, U.B.; Schuster, B. Liposome fusion on proteinaceous S-layer lattices triggered via β-diketone ligand–europium(III) complex formation. Soft Matter 2011, 7, 5514–5518. [Google Scholar] [CrossRef]
- Schrems, A.; Kibrom, A.; Küpcü, S.; Kiene, E.; Sleytr, U.B.; Schuster, B. Bilayer lipid membrane formation on a chemically modified S-layer lattice. Langmuir 2011, 27, 3731–3738. [Google Scholar] [CrossRef] [PubMed]
- Johannsmann, D. Viscoelastic, mechanical, and dielectric measurements on complex samples with the quartz crystal microbalance. Phys. Chem. Chem. Phys. 2008, 10, 4516–4534. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.K.; Wimley, W.C.; Searson, P.C.; Hristova, K.; Merzlyakov, M. Characterization of antimicrobial peptide activity by electrochemical impedance spectroscopy. Biochim. Biophys. Acta 2008, 1778, 2430–2436. [Google Scholar] [CrossRef] [PubMed]
- Bhakdi, S.; Tranum-Jensen, J. α-Toxin of staphylococcus aureus. Microbiol. Rev. 1991, 55, 733–751. [Google Scholar] [PubMed]
- Song, L.; Hobaugh, M.R.; Shustak, C.; Cheley, S.; Bayley, H.; Gouaux, J.E. Structure of staphylococcal α-hemolysin, a heptameric transmembrane pore. Science 1996, 274, 1859–1865. [Google Scholar] [CrossRef] [PubMed]
- Schuster, B.; Pum, D.; Braha, O.; Bayley, H.; Sleytr, U.B. Self-assembled α-hemolysin pores in an S-layer-supported lipid bilayer. Biochim. Biophys. Acta 1998, 1370, 280–288. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, S.; Wallace, B.A.; Morrow, J.S.; Veatch, W.R. Conformation of the gramicidin A transmembrane channel. J. Mol. Biol. 1980, 143, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Janshoff, A.; Steinem, C. Transport across artificial membranes—An analytical perspective. Anal. Bioanal. Chem. 2007, 385, 433–435. [Google Scholar] [CrossRef]
- Schrems, A.; Larisch, V.D.; Sleytr, U.B.; Hohenegger, M.; Lohner, K.; Schuster, B. Insertion of an anionic analogue of the antimicrobial peptide PGLa in lipid architectures including S-layer supported lipid bilayers. Curr. Nanosci. 2013, 9, 262–270. [Google Scholar] [CrossRef]
- Göbel, C.; Schuster, B.; Baurecht, D.; Sleytr, U.B.; Pum, D. S-layer templated bioinspired synthesis of silica. Colloids Surf. B Biointerfaces 2010, 75, 565–572. [Google Scholar] [CrossRef] [PubMed]
- Schuster, B.; Sleytr, U.B. Tailor-made crystalline structures of truncated S-layer proteins on heteropolysaccharides. Soft Matter 2009, 5, 334–341. [Google Scholar] [CrossRef]
- Gufler, P.; Pum, D.; Sleytr, U.B.; Schuster, B. Highly robust lipid membranes on crystalline S-layer supports investigated by electrochemical impedance spectroscopy. Biochim. Biophys. Acta 2004, 1661, 154–165. [Google Scholar] [CrossRef] [PubMed]
- Yuldasheva, L.; Carvalho, E.B.; Catanho, J.A.; Krasilnikov, O.V. Cholesterol-dependent hemolytic activity of Passiflora quadrangularis leaves. Braz. J. Med. Biol. Res. 2005, 38, 1061–1070. [Google Scholar] [CrossRef] [PubMed]
- Ashrafuzzaman, M.; Andersen, O.; McElhaney, R. The antimicrobial peptide gramicidin S permeabilizes phoshoplipid bilayer membranes without forming discrete ion. Biochim. Biophys. Acta 2008, 1778, 2814–2822. [Google Scholar] [CrossRef] [PubMed]
- Haluska, C.K.; Riske, K.A.; Marchi-Artzner, V.; Lehn, J.M.; Lipowsky, R.; Dimova, R. Time scales of membrane fusion revealed by direct imaging of vesicle fusion with high temporal resolution. Proc. Natl. Acad. Sci. USA 2006, 103, 15841–15846. [Google Scholar] [CrossRef] [PubMed]
- Reimhult, E.; Höök, F.; Kasemo, B. Intact vesicle adsorption and supported biomembrane formation from vesicles in solution: Influence of surface chemistry, vesicle size, temperature, and osmotic pressure. Langmuir 2003, 19, 1681–1691. [Google Scholar] [CrossRef]
- He, L.; Robertson, J.W.F.; Li, J.; Kärcher, I.; Schiller, S.M.; Knoll, W.; Naumann, R. Tethered bilayer lipid membranes based on monolayers of thiolipids mixed with a complementary dilution molecule: 1. Incorporation of channel peptides. Langmuir 2005, 21, 11666–11672. [Google Scholar] [CrossRef] [PubMed]
- Reimhult, E.; Kasemo, B.; Höök, F. Rupture pathway of phosphatidylcholic liposomes on silicon dioxide. Int. J. Mol. Sci. 2009, 10, 1683–1696. [Google Scholar] [CrossRef] [PubMed]
- John, K.; Schreiber, S.; Kubelt, J.; Herrmann, A.; Müller, P. Transbilayer movement of phospholipids at the main phase transition of lipid membranes: Implications for rapid flip-flop in biological membranes. Biophys. J. 2002, 83, 3315–3323. [Google Scholar] [CrossRef] [PubMed]
- Konradi, R.; Textor, M.; Reimhult, E. Using complementary acoustic and optical techniques for quantitative monitoring of biomolecular adsorption at interfaces. Biosensors 2012, 2, 341–376. [Google Scholar] [CrossRef] [PubMed]
- Kenausis, G.L.; Vörös, J.; Elbert, D.L.; Huang, N.; Hofer, R.; Ruiz-Taylor, L.; Textor, M.; Hubbell, J.A.; Spencer, N.D. Poly(l-lysine)-g-poly(ethylene glycol) layers on metal oxide surfaces: Attachment mechanism and effects of polymer architecture on resistance to protein adsorption. J. Phys. Chem. B 2000, 104, 3298–3309. [Google Scholar] [CrossRef]
- Huang, N.P.; Michel, R.; Vörös, J.; Textor, M.; Hofer, R.; Rossi, A.; Elbert, D.L.; Hubbell, J.A.; Spencer, N.D. Poly(l-lysine)-g-poly(ethylene glycol) layers on metal oxide surfaces: Surface-analytical characterization and resistance to serum and fibrinogen adsorption. Langmuir 2001, 17, 489–498. [Google Scholar] [CrossRef]
- Bakas, L.; Ostolaza, H.; Goni, F. Reversible adsorption and nonreversible insertion of Escherichia coli α-hemolysin into lipid bilayers. Biophys. J. 1996, 71, 1869–1876. [Google Scholar] [CrossRef] [PubMed]
- Nezil, F.; Bloom, M. Combined influence of cholesterol and synthetic amphiphilic peptides upon bilayer thickness in model membranes. Biophys. J. 1992, 61, 1176–1182. [Google Scholar] [CrossRef] [PubMed]
- Schuster, B.; Sleytr, U.B. Single channel recordings of α-hemolysin reconstituted in S-layer stabilized lipid bilayers. Bioelectrochem. 2002, 55, 5–7. [Google Scholar] [CrossRef]
- Glazier, S.A.; Vanderah, D.J.; Plant, A.L.; Bayley, H.; Valincius, G.; Kasianowicz, J.J. Reconstitution of the pore-forming toxin α-hemolysin in phospholipid/18-octadecyl-1-thiahexa (ethylene oxide) and phospholipid/n-octadecanethiol supported bilayer membranes. Langmuir 2000, 16, 10428–10435. [Google Scholar] [CrossRef]
- Sleytr, U.B.; Sara, M.; Küpcü, S.; Messner, P. Structural and chemical characterization of S-layers of selected strains of Bacillus stearothermophilus and Desulfotomaculum nigrificans. Arch. Microbiol. 1986, 146, 1–19. [Google Scholar] [CrossRef]
- Marchi-Artzner, V.; Brienne, M.J.; Gulik-Krzywicki, T.; Dedieu, J.C.; Lehn, J.M. Selective complexation and transport of europium ions at the interface of vesicles. Chem. Eur. J. 2004, 10, 2342–2350. [Google Scholar] [CrossRef] [PubMed]
- Vallejo, A.E.; Gervasi, C.A. On the use of impedance spectroscopy for studying bilayer lipid membranes. In Advances in Planar Lipid Bilayers and Liposomes; Tien, H.T., Ottova-Leitmannova, A., Eds.; Academic Press: Waltham, MA, USA, 2006; Volume 5, pp. 331–351. [Google Scholar]
- Terrettaz, S.; Stora, T.; Duschl, C.; Vogel, H. Protein binding to supported lipid membranes: Investigation of the cholera toxin-ganglioside interaction by simultaneous impedance spectroscopy and surface plasmon resonance. Langmuir 1993, 9, 1361–1369. [Google Scholar] [CrossRef]
- Naumann, R.; Walz, D.; Schiller, S.; Knoll, W. Kinetics of valinomycin-mediated K+-ion transport through tethered bilayer lipid membranes. J. Electroanal. Chem. 2003, 50, 241–252. [Google Scholar] [CrossRef]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Damiati, S.; Schrems, A.; Sinner, E.-K.; Sleytr, U.B.; Schuster, B. Probing Peptide and Protein Insertion in a Biomimetic S-Layer Supported Lipid Membrane Platform. Int. J. Mol. Sci. 2015, 16, 2824-2838. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms16022824
Damiati S, Schrems A, Sinner E-K, Sleytr UB, Schuster B. Probing Peptide and Protein Insertion in a Biomimetic S-Layer Supported Lipid Membrane Platform. International Journal of Molecular Sciences. 2015; 16(2):2824-2838. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms16022824
Chicago/Turabian StyleDamiati, Samar, Angelika Schrems, Eva-Kathrin Sinner, Uwe B. Sleytr, and Bernhard Schuster. 2015. "Probing Peptide and Protein Insertion in a Biomimetic S-Layer Supported Lipid Membrane Platform" International Journal of Molecular Sciences 16, no. 2: 2824-2838. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms16022824