Favourable IFNL3 Genotypes Are Associated with Spontaneous Clearance and Are Differentially Distributed in Aboriginals in Canadian HIV-Hepatitis C Co-Infected Individuals
Abstract
:1. Introduction
2. Results
2.1. Clearance and IFNL3
| Variables | Study Population n = 538 | CCC n = 1176 |
|---|---|---|
| Median follow-up time, years (IQR) | 3.2 (1.7–4.6) | 3.0 (1–4.4) |
| Mean age at baseline, years (SD) | 44 (8.2) | 45 (8.6) |
| Male, n (%) | 368 (68) | 864 (74) |
| Ethnicity, n (%) | ||
| White | 418 (78) | 891 (77) |
| Black | 15 (3) | 45 (4) |
| Aboriginal | 85 (16) | 181 (16) |
| Other | 15 (3) | 44 (4) |
| Injection drug use ever, n (%) | 472 (87) | 944 (81) |
| Median HCV duration, years (IQR) | 19 (11–25) | 18 (10–26) |
| HCV RNA positive at first available test | 481 (90) | 889 (76) |
| HCV genotype 1, n (%) | 304 (74) a | 683 (73) b |
| Median CD4 counts, cells/μL (IQR) | 365 (230–530) | 420 (270–604) |
| On HIV therapy | 415 (77) | 957 (81) |
| Variables | Spontaneous Clearers n = 79 | Chronically Infected n = 462 | Univariate HR (95% CI) |
|---|---|---|---|
| Aboriginal | 18 (23%) | 67 (15%) | 1.91 (1.12, 3.25) a |
| Female | 32 (41%) | 142 (31%) | 1.62 (1.02, 2.57) |
| HCV genotype, n (%) | |||
| 1 | 11 (52%) | 293 (75%) | 0.56 (0.29, 1.10) b |
| 2 | 1 (5%) | 17 (4%) | |
| 3 | 9 (43%) | 67 (17%) | |
| 4 | 0 | 12 (3%) | |
| IFNL3 genotypes | |||
| rs12979860 | |||
| CC, n (%) | 53 (75%) | 180 (43%) | 3.89 (2.28, 6.63) c |
| CT, n (%) | 15 (21%) | 186 (44%) | |
| TT, n (%) | 3 (4%) | 57 (13%) | |
| rs8099917 | |||
| TT, n (%) | 68 (88%) | 256 (60%) | 4.65 (2.32, 9.32) |
| GT, n (%) | 9 (12%) | 138 (33%) | |
| GG, n (%) | 0 (0) | 30 (7%) | |
| rs8103142 | |||
| TT, n (%) | 59 (78%) | 186 (45%) | 4.23 (2.46, 7.28) |
| CT, n (%) | 15 (20%) | 182 (44%) | |
| CC, n (%) | 2 (2%) | 50 (12%) | |
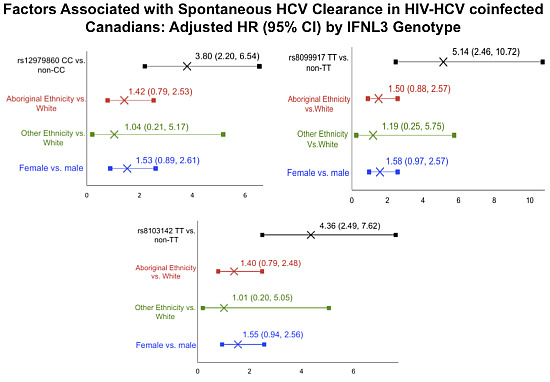
| Characteristic | Adjusted HR (95% CI) by IFNL3 Genotype | ||
|---|---|---|---|
| rs12979860 CC | rs8099917 TT | rs8103142 TT | |
| 3.80 (2.20, 6.54) | 5.14 (2.46, 10.72) | 4.36 (2.49,7.62) | |
| Aboriginal Ethnicity vs. White | 1.42 (0.79, 2.53) | 1.50 (0.88, 2.57) | 1.40 (0.79, 2.48) |
| Other Ethnicity vs. White | 1.04 (0.21, 5.17) | 1.19(0.25, 5.75) | 1.01 (0.20, 5.05) |
| Female | 1.53 (0.89, 2.61) | 1.58 (0.97, 2.57) | 1.55 (0.94, 2.56) |
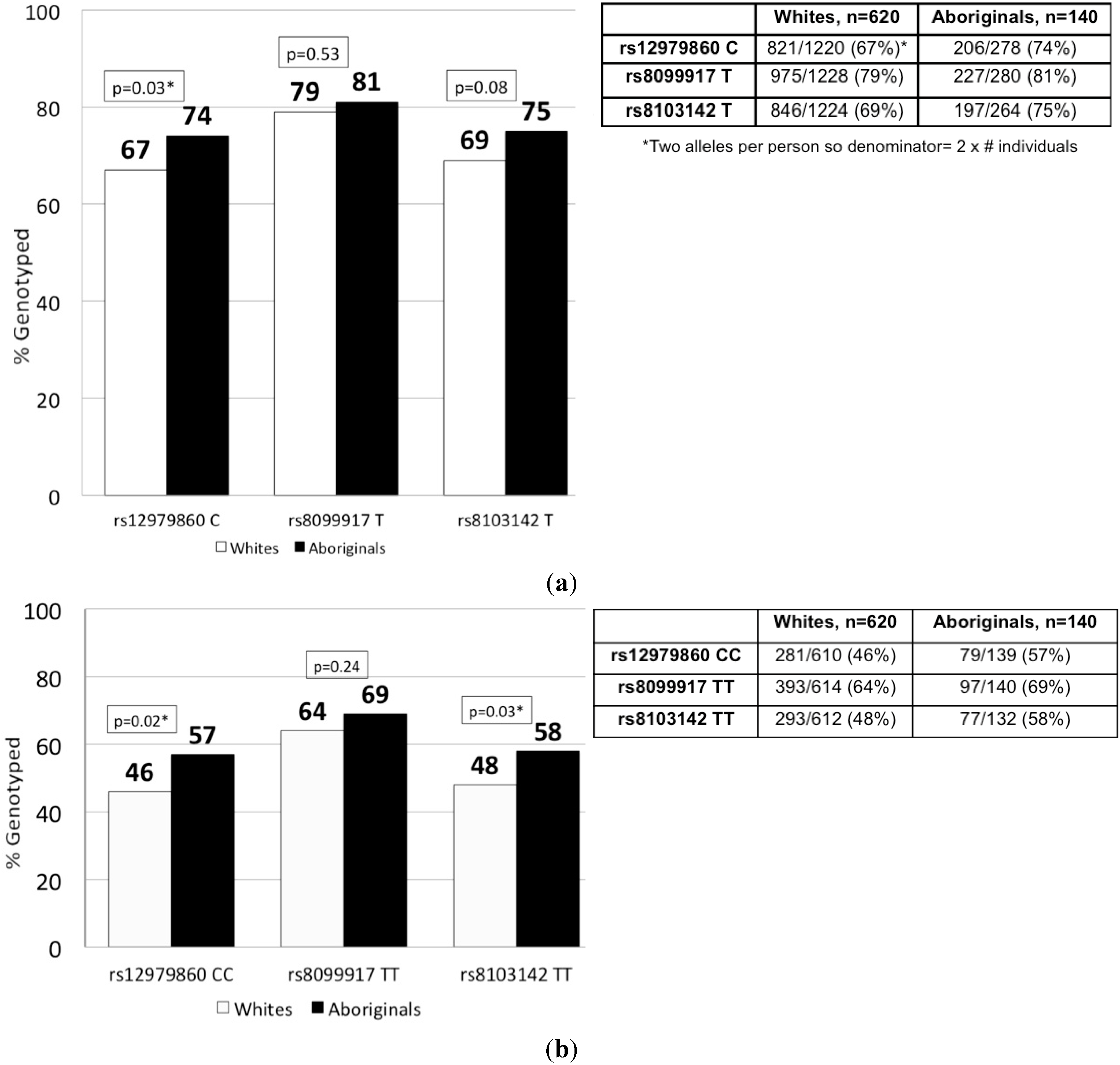
2.2. IFNL3 Distribution in Aboriginals and Whites

3. Discussion
4. Materials and Methods
4.1. Source Population
4.2. Study Population and Covariates

4.3. IFNL3 SNP Genotyping
4.4. Statistical Analysis
Acknowledgments
Author Contributions
Appendix
Conflicts of Interest
References
- Thomas, D.L.; Thio, C.L.; Martin, M.P.; Qi, Y.; Ge, D.; O’Huigin, C.; Kidd, J.; Kidd, K.; Khakoo, S.I.; Alexander, G.; et al. Genetic variation in IL28B and spontaneous clearance of hepatitis c virus. Nature 2009, 461, 798–801. [Google Scholar] [CrossRef]
- Grebely, J.; Petoumenos, K.; Hellard, M.; Matthews, G.V.; Suppiah, V.; Applegate, T.; Yeung, B.; Marks, P.; Rawlinson, W.; Lloyd, A.R.; et al. Potential role for interleukin-28b genotype in treatment decision-making in recent hepatitis c virus infection. Hepatology 2010, 52, 1216–1224. [Google Scholar] [CrossRef]
- Montes-Cano, M.A.; Garcia-Lozano, J.R.; Abad-Molina, C.; Romero-Gomez, M.; Barroso, N.; Aguilar-Reina, J.; Nunez-Roldan, A.; Gonzalez-Escribano, M.F. Interleukin-28B genetic variants and hepatitis virus infection by different viral genotypes. Hepatology 2010, 52, 33–37. [Google Scholar] [CrossRef] [PubMed]
- Rauch, A.; Kutalik, Z.; Descombes, P.; Cai, T.; Di Iulio, J.; Mueller, T.; Bochud, M.; Battegay, M.; Bernasconi, E.; Borovicka, J.; et al. Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: A genome-wide association study. Gastroenterology 2010, 138, 1338–1345. [Google Scholar] [CrossRef]
- Mizukoshi, E.; Eisenbach, C.; Edlin, B.R.; Newton, K.P.; Raghuraman, S.; Weiler-Normann, C.; Tobler, L.H.; Busch, M.P.; Carrington, M.; McKeating, J.A.; et al. Hepatitis c virus (HCV)-specific immune responses of long-term injection drug users frequently exposed to hcv. J. Infect. Dis. 2008, 198, 203–212. [Google Scholar] [CrossRef]
- Wang, C.C.; Krantz, E.; Klarquist, J.; Krows, M.; McBride, L.; Scott, E.P.; Shaw-Stiffel, T.; Weston, S.J.; Thiede, H.; Wald, A.; et al. Acute hepatitis C in a contemporary us cohort: Modes of acquisition and factors influencing viral clearance. J. Infect. Dis. 2007, 196, 1474–1482. [Google Scholar] [CrossRef]
- Kamal, S.M.; Kassim, S.K.; Ahmed, A.I.; Mahmoud, S.; Bahnasy, K.A.; Hafez, T.A.; Aziz, I.A.; Fathelbab, I.F.; Mansour, H.M. Host and viral determinants of the outcome of exposure to HCV infection genotype 4: A large longitudinal study. Am. J. Gastroenterol. 2014, 109, 199–211. [Google Scholar] [PubMed]
- Rao, H.Y.; Sun, D.G.; Jiang, D.; Yang, R.F.; Guo, F.; Wang, J.H.; Liu, F.; Zhang, H.Y.; Zhang, H.H.; Du, S.C.; et al. IL28B genetic variants and gender are associated with spontaneous clearance of hepatitis c virus infection. J. Viral Hepat. 2012, 19, 173–181. [Google Scholar] [CrossRef] [Green Version]
- Dawood, M.; Smart, G.; Wood, M.; Wu, H.X.; Paton, S.; Wu, J. Hepatitis c virus infection among first nation and non-first nation people in Manitoba, Canada: A public health laboratory study. Can. J. Microbiol. 2006, 52, 999–1005. [Google Scholar] [CrossRef]
- Grebely, J.; Raffa, J.D.; Lai, C.; Krajden, M.; Conway, B.; Tyndall, M.W. Factors associated with spontaneous clearance of hepatitis c virus among illicit drug users. Can. J. Gastroenterol. 2007, 21, 447–451. [Google Scholar] [PubMed]
- Kim, A.Y.; Chung, R.T. Coinfection with HIV-1 and HCV—A one-two punch. Gastroenterology 2009, 137, 795–814. [Google Scholar] [CrossRef] [PubMed]
- Mosbruger, T.L.; Duggal, P.; Goedert, J.J.; Kirk, G.D.; Hoots, W.K.; Tobler, L.H.; Busch, M.; Peters, M.G.; Rosen, H.R.; Thomas, D.L.; et al. Large-scale candidate gene analysis of spontaneous clearance of hepatitis c virus. J. Infect. Dis. 2010, 201, 1371–1380. [Google Scholar] [CrossRef]
- Romero-Gomez, M.; Eslam, M.; Ruiz, A.; Maraver, M. Genes and hepatitis C: Susceptibility, fibrosis progression and response to treatment. Liver Int. 2011, 31, 443–460. [Google Scholar] [CrossRef] [PubMed]
- Aborsangaya, K.B.; Dembinski, I.; Khatkar, S.; Alphonse, M.P.; Nickerson, P.; Rempel, J.D. Impact of aboriginal ethnicity on HCV core-induced IL-10 synthesis: Interaction with IL-10 gene polymorphisms. Hepatology 2007, 45, 623–630. [Google Scholar] [CrossRef] [PubMed]
- Rempel, J.D.; Hawkins, K.; Lande, E.; Nickerson, P. The potential influence of kir cluster profiles on disease patterns of Canadian aboriginals and other Indigenous peoples of the americas. Eur. J. Hum. Genet. 2011, 19, 1276–1280. [Google Scholar] [CrossRef] [PubMed]
- Tillmann, H.L.; Thompson, A.J.; Patel, K.; Wiese, M.; Tenckhoff, H.; Nischalke, H.D.; Lokhnygina, Y.; Kullig, U.; Gobel, U.; Capka, E.; et al. A polymorphism near IL28B is associated with spontaneous clearance of acute hepatitis C virus and jaundice. Gastroenterology 2010, 139, 1586–1592. [Google Scholar] [CrossRef]
- Duggal, P.; Thio, C.L.; Wojcik, G.L.; Goedert, J.J.; Mangia, A.; Latanich, R.; Kim, A.Y.; Lauer, G.M.; Chung, R.T.; Peters, M.G.; et al. Genome-wide association study of spontaneous resolution of hepatitis C virus infection: Data from multiple cohorts. Ann. Intern. Med. 2013, 158, 235–245. [Google Scholar] [CrossRef]
- Ge, D.; Fellay, J.; Thompson, A.J.; Simon, J.S.; Shianna, K.V.; Urban, T.J.; Heinzen, E.L.; Qiu, P.; Bertelsen, A.H.; Muir, A.J.; et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature 2009, 461, 399–401. [Google Scholar] [CrossRef]
- Tanaka, Y.; Nishida, N.; Sugiyama, M.; Kurosaki, M.; Matsuura, K.; Sakamoto, N.; Nakagawa, M.; Korenaga, M.; Hino, K.; Hige, S.; et al. Genome-wide association of IL28B with response to pegylated interferon-α and ribavirin therapy for chronic hepatitis C. Nat. Genet. 2009, 41, 1105–1109. [Google Scholar] [CrossRef]
- Indolfi, G.; Mangone, G.; Calvo, P.L.; Bartolini, E.; Regoli, M.; Serranti, D.; Calitri, C.; Tovo, P.A.; de Martino, M.; Azzari, C.; et al. Interleukin 28B rs12979860 single-nucleotide polymorphism predicts spontaneous clearance of hepatitis C virus in children. J. Pediatr. Gastroenterol. Nutr. 2014, 58, 666–668. [Google Scholar] [CrossRef]
- Indolfi, G.; Sambrotta, M.; Moriondo, M.; Azzari, C.; Resti, M. Genetic variation in interleukin-28b locus is associated with spontaneous clearance of hcv in children with non-1 viral genotype infection. Hepatology 2011, 54, 1490–1491. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Extremera, A.; Munoz-Gamez, J.A.; Salmeron-Ruiz, M.A.; de Rueda, P.M.; Quiles-Perez, R.; Gila-Medina, A.; Casado, J.; Belen Martin, A.; Sanjuan-Nunez, L.; Carazo, A.; et al. Genetic variation in interleukin 28B with respect to vertical transmission of hepatitis C virus and spontaneous clearance in hcv-infected children. Hepatology 2011, 53, 1830–1838. [Google Scholar] [CrossRef]
- Prokunina-Olsson, L.; Muchmore, B.; Tang, W.; Pfeiffer, R.M.; Park, H.; Dickensheets, H.; Hergott, D.; Porter-Gill, P.; Mumy, A.; Kohaar, I.; et al. A variant upstream of IFNL3 (IL28B) creating a new interferon gene ifnl4 is associated with impaired clearance of hepatitis C virus. Nat. Genet. 2013, 45, 164–171. [Google Scholar] [CrossRef]
- Aka, P.V.; Kuniholm, M.H.; Pfeiffer, R.M.; Wang, A.S.; Tang, W.; Chen, S.; Astemborski, J.; Plankey, M.; Villacres, M.C.; Peters, M.G.; et al. Association of the IFNL4-Δ allele with impaired spontaneous clearance of hepatitis C virus. J. Infect. Dis. 2014, 209, 350–354. [Google Scholar] [CrossRef]
- Bibert, S.; Roger, T.; Calandra, T.; Bochud, M.; Cerny, A.; Semmo, N.; Duong, F.H.; Gerlach, T.; Malinverni, R.; Moradpour, D.; et al. IL28B expression depends on a novel TT/-G polymorphism which improves HCV clearance prediction. J. Exp. Med. 2013, 210, 1109–1116. [Google Scholar] [CrossRef]
- Clark, P.J.; Thompson, A.J. Host genomics and HCV treatment response. J. Gastroenterol. Hepatol. 2012, 27, 212–222. [Google Scholar] [CrossRef] [PubMed]
- De Castellarnau, M.; Aparicio, E.; Parera, M.; Franco, S.; Tural, C.; Clotet, B.; Martinez, M.A. Deciphering the interleukin 28B variants that better predict response to pegylated interferon-α and ribavirin therapy in HCV/HIV-1 coinfected patients. PLoS One 2012, 7, e31016. [Google Scholar] [CrossRef] [PubMed]
- Bellanti, F.; Vendemiale, G.; Altomare, E.; Serviddio, G. The impact of interferon lambda 3 gene polymorphism on natural course and treatment of hepatitis C. Clin. Dev. Immunol. 2012, 2012. [Google Scholar] [CrossRef]
- Poordad, F.; Bronowicki, J.P.; Gordon, S.C.; Zeuzem, S.; Jacobson, I.M.; Sulkowski, M.S.; Poynard, T.; Morgan, T.R.; Molony, C.; Pedicone, L.D.; et al. Factors that predict response of patients with hepatitis C virus infection to boceprevir. Gastroenterology 2012, 143, 608–618, e601–e605. [Google Scholar] [CrossRef]
- Akuta, N.; Suzuki, F.; Hirakawa, M.; Kawamura, Y.; Yatsuji, H.; Sezaki, H.; Suzuki, Y.; Hosaka, T.; Kobayashi, M.; Kobayashi, M.; et al. Amino acid substitution in hepatitis c virus core region and genetic variation near the interleukin 28B gene predict viral response to telaprevir with peginterferon and ribavirin. Hepatology 2010, 52, 421–429. [Google Scholar] [CrossRef]
- Chu, T.W.; Kulkarni, R.; Gane, E.J.; Roberts, S.K.; Stedman, C.; Angus, P.W.; Ritchie, B.; Lu, X.Y.; Ipe, D.; Lopatin, U.; et al. Effect of IL28B genotype on early viral kinetics during interferon-free treatment of patients with chronic hepatitis C. Gastroenterology 2012, 142, 790–795. [Google Scholar] [CrossRef]
- Wu, H.X.; Wu, J.; Wong, T.; Andonov, A.; Li, Q.; Dinner, K.; Donaldson, T.; Paton, S. Incidence and risk factors for newly acquired hepatitis C virus infection among aboriginal vs. non-aboriginal canadians in six regions, 1999–2004. Eur. J. Clin. Microbiol. Infect. Dis. 2007, 26, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Rempel, J.D.; Uhanova, J. Hepatitis c virus in american indian/alaskan native and aboriginal peoples of north america. Viruses 2012, 4, 3912–3931. [Google Scholar] [CrossRef] [PubMed]
- Statistics, C. 2011 National Household Survey: Aboriginal Peoples in Canada: First Nations People, Métis and Inuit. Available online: http://www.statcan.gc.ca/daily-quotidien/130508/dq130508a-eng.htm?HPA (accessed on 6 June 2014).
- Lelutiu-Weinberger, C.; Pouget, E.R.; Des Jarlais, D.D.; Cooper, H.L.; Scheinmann, R.; Stern, R.; Strauss, S.M.; Hagan, H. A meta-analysis of the hepatitis c virus distribution in diverse racial/ethnic drug injector groups. Soc. Sci. Med. 2009, 68, 579–590. [Google Scholar] [CrossRef] [PubMed]
- Minuk, G.Y.; Uhanova, J. Viral hepatitis in the canadian inuit and first nations populations. Can. J. Gastroenterol. 2003, 17, 707–712. [Google Scholar] [PubMed]
- Minuk, G.Y.; Zhang, M.; Wong, S.G.; Uhanova, J.; Bernstein, C.N.; Martin, B.; Dawood, M.R.; Vardy, L.; Giulvi, A. Viral hepatitis in a canadian first nations community. Can. J. Gastroenterol. 2003, 17, 593–596. [Google Scholar] [PubMed]
- Rempel, J.D.; Aborsangaya, K.B.; Alphonse, M.P.; Minuk, G.Y. The influence of north american aboriginal ethnicity on pro-inflammatory and anti-inflammatory cytokine responses to IFN-α. J. Viral Hepat. 2009, 16, 292–297. [Google Scholar] [CrossRef] [PubMed]
- Cooper, C.L.; Bailey, R.J.; Bain, V.G.; Anderson, F.; Yoshida, E.M.; Krajden, M.; Marotta, P. Outcomes of peginterferon α-2A and ribavirin hepatitis c therapy in aboriginal canadians. Can. J. Gastroenterol. 2008, 22, 677–680. [Google Scholar] [PubMed]
- Minuk, G.Y.; O’Brien, M.; Hawkins, K.; Emokpare, D.; McHattie, J.; Harris, P.; Worobetz, L.; Doucette, K.; Kaita, K.; Wong, S.; et al. Treatment of chronic hepatitis c in a canadian aboriginal population: Results from the prairie study. Can. J. Gastroenterol. 2013, 27, 707–710. [Google Scholar] [PubMed]
- Wang, S.; Lewis, C.M.; Jakobsson, M.; Ramachandran, S.; Ray, N.; Bedoya, G.; Rojas, W.; Parra, M.V.; Molina, J.A.; Gallo, C.; et al. Genetic variation and population structure in native americans. PLoS Genet. 2007, 3, e185. [Google Scholar] [CrossRef] [PubMed]
- Reich, D.; Patterson, N.; Campbell, D.; Tandon, A.; Mazieres, S.; Ray, N.; Parra, M.V.; Rojas, W.; Duque, C.; Mesa, N.; et al. Reconstructing native american population history. Nature 2012, 488, 370–374. [Google Scholar] [CrossRef] [PubMed]
- Clausen, L.N.; Weis, N.; Astvad, K.; Schonning, K.; Fenger, M.; Krarup, H.; Bukh, J.; Benfield, T. Interleukin-28B polymorphisms are associated with hepatitis C virus clearance and viral load in a HIV-1-infected cohort. J. Viral Hepat. 2011, 18, e66–e74. [Google Scholar] [CrossRef] [PubMed]
- Clausen, L.N.; Lundbo, L.F.; Benfield, T. Hepatitis C virus infection in the human immunodeficiency virus infected patient. World J. Gastroenterol. 2014, 20, 12132–12143. [Google Scholar] [CrossRef] [PubMed]
- Di Iulio, J.; Ciuffi, A.; Fitzmaurice, K.; Kelleher, D.; Rotger, M.; Fellay, J.; Martinez, R.; Pulit, S.; Furrer, H.; Gunthard, H.F.; et al. Estimating the net contribution of interleukin-28b variation to spontaneous hepatitis c virus clearance. Hepatology 2011, 53, 1446–1454. [Google Scholar] [CrossRef] [PubMed]
- Pedergnana, V.; Abdel-Hamid, M.; Guergnon, J.; Mohsen, A.; Le Fouler, L.; Theodorou, I.; Mohamed, M.K.; Fontanet, A.; Plancoulaine, S.; Abel, L. Analysis of IL28B variants in an egyptian population defines the 20 kilobases minimal region involved in spontaneous clearance of hepatitis c virus. PLoS One 2012, 7, e38578. [Google Scholar] [CrossRef] [PubMed]
- Balagopal, A.; Thomas, D.L.; Thio, C.L. IL28b and the control of hepatitis C virus infection. Gastroenterology 2010, 139, 1865–1876. [Google Scholar] [CrossRef] [PubMed]
- Suppiah, V.; Gaudieri, S.; Armstrong, N.J.; O’Connor, K.S.; Berg, T.; Weltman, M.; Abate, M.L.; Spengler, U.; Bassendine, M.; Dore, G.J.; et al. Il28b, HLA-C, and kir variants additively predict response to therapy in chronic hepatitis c virus infection in a european cohort: A cross-sectional study. PLoS Med. 2011, 8, e1001092. [Google Scholar] [CrossRef] [PubMed]
- The International HapMap Consortium. The international hapmap project. Nature 2003, 426, 789–796. [Google Scholar]
- Uhanova, J.; Tate, R.B.; Tataryn, D.J.; Minuk, G.Y. The epidemiology of hepatitis c in a canadian indigenous population. Can. J. Gastroenterol. 2013, 27, 336–340. [Google Scholar] [PubMed]
- Sugiyama, M.; Tanaka, Y.; Wakita, T.; Nakanishi, M.; Mizokami, M. Genetic variation of the IL-28B promoter affecting gene expression. PLoS One 2011, 6, e26620. [Google Scholar] [CrossRef] [PubMed]
- Urban, T.J.; Thompson, A.J.; Bradrick, S.S.; Fellay, J.; Schuppan, D.; Cronin, K.D.; Hong, L.; McKenzie, A.; Patel, K.; Shianna, K.V.; et al. IL28B genotype is associated with differential expression of intrahepatic interferon-stimulated genes in patients with chronic hepatitis C. Hepatology 2010, 52, 1888–1896. [Google Scholar] [CrossRef] [PubMed]
- Urban, T.; Charlton, M.R.; Goldstein, D.B. Introduction to the genetics and biology of interleukin-28B. Hepatology 2012, 56, 361–366. [Google Scholar] [CrossRef] [PubMed]
- Chinnaswamy, S. Genetic variants at the IFNL3 locus and their association with hepatitis C virus infections reveal novel insights into host-virus interactions. J. Interferon Cytokine Res. 2014, 34, 479–497. [Google Scholar] [CrossRef] [PubMed]
- Van den Berg, C.H.; Grady, B.P.; Schinkel, J.; van de Laar, T.; Molenkamp, R.; van Houdt, R.; Coutinho, R.A.; van Baarle, D.; Prins, M. Female sex and IL28B, a synergism for spontaneous viral clearance in hepatitis C virus (HCV) seroconverters from a community-based cohort. PLoS One 2011, 6, e27555. [Google Scholar] [CrossRef] [PubMed]
- Klein, M.B.; Saeed, S.; Yang, H.; Cohen, J.; Conway, B.; Cooper, C.; Cote, P.; Cox, J.; Gill, J.; Haase, D.; et al. Cohort profile: The canadian HIV-hepatitis c co-infection cohort study. Int. J. Epidemiol. 2010, 39, 1162–1169. [Google Scholar] [CrossRef] [PubMed]
- Stephens, M.; Donnelly, P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am. J. Hum. Genet. 2003, 73, 1162–1169. [Google Scholar] [CrossRef] [PubMed]
- Stephens, M.; Smith, N.J.; Donnelly, P. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 2001, 68, 978–989. [Google Scholar] [CrossRef] [PubMed]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef] [PubMed]
- White, I.R.; Royston, P.; Wood, A.M. Multiple imputation using chained equations: Issues and guidance for practice. Stat. Med. 2011, 30, 377–399. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moqueet, N.; Infante-Rivard, C.; Platt, R.W.; Young, J.; Cooper, C.; Hull, M.; Walmsley, S.; Klein, M.B.; Investigators, T.C.C.-I.S. Favourable IFNL3 Genotypes Are Associated with Spontaneous Clearance and Are Differentially Distributed in Aboriginals in Canadian HIV-Hepatitis C Co-Infected Individuals. Int. J. Mol. Sci. 2015, 16, 6496-6512. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms16036496
Moqueet N, Infante-Rivard C, Platt RW, Young J, Cooper C, Hull M, Walmsley S, Klein MB, Investigators TCC-IS. Favourable IFNL3 Genotypes Are Associated with Spontaneous Clearance and Are Differentially Distributed in Aboriginals in Canadian HIV-Hepatitis C Co-Infected Individuals. International Journal of Molecular Sciences. 2015; 16(3):6496-6512. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms16036496
Chicago/Turabian StyleMoqueet, Nasheed, Claire Infante-Rivard, Robert W. Platt, Jim Young, Curtis Cooper, Mark Hull, Sharon Walmsley, Marina B. Klein, and The Canadian Co-Infection Study Investigators. 2015. "Favourable IFNL3 Genotypes Are Associated with Spontaneous Clearance and Are Differentially Distributed in Aboriginals in Canadian HIV-Hepatitis C Co-Infected Individuals" International Journal of Molecular Sciences 16, no. 3: 6496-6512. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms16036496