Role of Intracellular Lipid Logistics in the Preferential Usage of Very Long Chain-Ceramides in Glucosylceramide
Abstract
:1. Introduction
2. Results
2.1. Preparation of CERS2 Mutants in HeLa Cells by TALEN
2.2. Differential Usage of VLC-Cer between GSLs and SM
2.3. Enrichment of De Novo Synthesized C16:0-Cer in the ER Did Not Affect the Preferential Usage of VLC-Cer in GlcCer
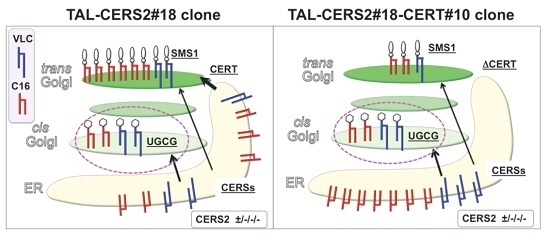
2.4. Reduction in the Proportion of VLC-GlcCer by Treatment with Brefeldin A and Overexpression of UGCG
3. Discussion
4. Materials and Methods
4.1. Cell Culture, Antibodies, and Reagents
4.2. Synthesis of TALEN Constructs
4.3. Subcloning of CERS2 and UGCG cDNAs in Retroviral Vectors and Their Retroviral Infection
4.4. Genome editing with TALENs
4.5. mRNA Analysis
4.6. Metabolic Labeling of Sphingolipids and TLC Analysis
4.7. Western Blot
4.8. Cell Growth
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Hanada, K.; Nishijima, M.; Kiso, M.; Hasegawa, A.; Fujita, S.; Ogawa, T.; Akamatsu, Y. Sphingolipids are essential for the growth of Chinese hamster ovary cells. Restoration of the growth of a mutant defective in sphingoid base biosynthesis by exogenous sphingolipids. J. Biol. Chem. 1992, 267, 23527–23533. [Google Scholar] [PubMed]
- Pinto, W.J.; Wells, G.W.; Lester, R.L. Characterization of enzymatic synthesis of sphingolipid long-chain bases in Saccharomyces cerevisiae: Mutant strains exhibiting long-chain-base auxotrophy are deficient in serine palmitoyltransferase activity. J. Bacteriol. 1992, 174, 2575–2581. [Google Scholar] [PubMed]
- Hakomori, S. Structure and function of glycosphingolipids and sphingolipids: Recollections and future trends. Biochim. Biophys. Acta 2008, 1780, 325–346. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Liu, Y.; Sullards, M.C.; Merrill, A.H., Jr. An introduction to sphingolipid metabolism and analysis by new technologies. Neuromol. Med. 2010, 12, 306–319. [Google Scholar] [CrossRef] [PubMed]
- Wennekes, T.; van den Berg, R.J.; Boot, R.G.; van der Marel, G.A.; Overkleeft, H.S.; Aerts, J.M. Glycosphingolipids—Nature, function, and pharmacological modulation. Angew. Chem. Int. Ed. Engl. 2009, 48, 8848–8869. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, K.; Ohmi, Y.; Ohkawa, Y.; Tokuda, N.; Kondo, Y.; Tajima, O.; Furukawa, K. Regulatory mechanisms of nervous systems with glycosphingolipids. Neurochem. Res. 2011, 36, 1578–1586. [Google Scholar] [CrossRef] [PubMed]
- Proia, R. Glycosphingolipid functions: Insights from engineered mouse models. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2003, 538, 879–883. [Google Scholar] [CrossRef] [PubMed]
- Halter, D.; Neumann, S.; van Dijk, S.M.; Wolthoorn, J.; de Mazière, A.M.; Vieira, O.V.; Mattjus, P.; Klumperman, J.; van Meer, G.; Sprong, H. Pre- and post-Golgi translocation of glucosylceramide in glycosphingolipid synthesis. J. Cell Biol. 2007, 179, 101–115. [Google Scholar] [CrossRef] [PubMed]
- Hanada, K.; Kumagai, K.; Yasuda, S.; Miura, Y.; Kawano, M.; Fukasawa, M.; Nishijima, M. Molecular machinery for non-vesicular trafficking of ceramide. Nature 2003, 426, 803–809. [Google Scholar] [CrossRef] [PubMed]
- Yamaji, T.; Hanada, K. Sphingolipid metabolism and interorganellar transport: Localization of sphingolipid enzymes and lipid transfer proteins. Traffic 2015, 60, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Giussani, P.; Brioschi, L.; Bassi, R.; Riboni, L.; Viani, P. Phosphatidylinositol 3-kinase/AKT pathway regulates the endoplasmic reticulum to golgi traffic of ceramide in glioma cells: A link between lipid signaling pathways involved in the control of cell survival. J. Biol. Chem. 2009, 284, 5088–5096. [Google Scholar] [CrossRef] [PubMed]
- Gillon, A.; Latham, C.F.; Miller, E.A. Vesicle-mediated ER export of proteins and lipids. Biochim. Biophys. Acta 2012, 1821, 1040–1049. [Google Scholar] [CrossRef] [PubMed]
- Pewzner-Jung, Y.; Ben-Dor, S.; Futerman, A.H. When do Lasses (longevity assurance genes) become CerS (ceramide synthases)? Insights into the regulation of ceramide synthesis. J. Biol. Chem. 2006, 281, 25001–25005. [Google Scholar] [CrossRef] [PubMed]
- Levy, M.; Futerman, A.H. Mammalian ceramide synthases. IUBMB Life 2010, 62, 347–356. [Google Scholar] [CrossRef] [PubMed]
- Venkataraman, K.; Riebeling, C.; Bodennec, J.; Riezman, H.; Allegood, J.C.; Sullards, M.C.; Merrill, A.H., Jr.; Futerman, A.H. Upstream of growth and differentiation factor 1 (uog1), a mammalian homolog of the yeast longevity assurance gene 1 (LAG1), regulates N-stearoyl-sphinganine (C18-(dihydro)ceramide) synthesis in a fumonisin B1-independent manner in mammalian cells. J. Biol. Chem. 2002, 277, 35642–35649. [Google Scholar] [CrossRef] [PubMed]
- Riebeling, C.; Allegood, J.C.; Wang, E.; Merrill, A.H., Jr.; Futerman, A.H. Two mammalian longevity assurance gene (LAG1) family members, trh1 and trh4, regulate dihydroceramide synthesis using different fatty acyl-CoA donors. J. Biol. Chem. 2003, 278, 43452–43459. [Google Scholar] [CrossRef] [PubMed]
- Laviad, E.L.; Albee, L.; Pankova-Kholmyansky, I.; Epstein, S.; Park, H.; Merrill, A.H., Jr.; Futerman, A.H. Characterization of ceramide synthase 2: Tissue distribution, substrate specificity, and inhibition by sphingosine 1-phosphate. J. Biol. Chem. 2008, 283, 5677–5684. [Google Scholar] [CrossRef] [PubMed]
- Mizutani, Y.; Kihara, A.; Igarashi, Y. Mammalian Lass6 and its related family members regulate synthesis of specific ceramides. Biochem. J. 2005, 390, 263–271. [Google Scholar] [CrossRef] [PubMed]
- Mizutani, Y.; Kihara, A.; Igarashi, Y. LASS3 (longevityassurance homologue 3) is a mainly testis-specific (dihydro)ceramide synthase with relatively broad substrate specificity. Biochem. J. 2006, 398, 531–538. [Google Scholar] [CrossRef] [PubMed]
- Wegner, M.S.; Schiffmann, S.; Parnham, M.J.; Geisslinger, G.; Grösch, S. The enigma of ceramide synthase regulation in mammalian cells. Prog. Lipid Res. 2016, 63, 93–119. [Google Scholar] [CrossRef] [PubMed]
- Pewzner-Jung, Y.; Brenner, O.; Braun, S.; Laviad, E.L.; Ben-Dor, S.; Feldmesser, E.; Horn-Saban, S.; Amann-Zalcenstein, D.; Raanan, C.; Berkutzki, T.; et al. A critical role for ceramide synthase 2 in liver homeostasis II. Insights into molecular changes leading to hepatopathy. J. Biol. Chem. 2010, 285, 10911–10923. [Google Scholar] [CrossRef] [PubMed]
- Park, W.J.; Park, J.W. The effect of altered sphingolipid acyl chain length on various disease models. Biol. Chem. 2015, 396, 693–705. [Google Scholar] [CrossRef] [PubMed]
- Iwabuchi, K.; Prinetti, A.; Sonnino, S.; Mauri, L.; Kobayashi, T.; Ishii, K.; Kaga, N.; Murayama, K.; Kurihara, H.; Nakayama, H.; et al. Involvement of very long fatty acid-containing lactosylceramide in lactosylceramide-mediated superoxide generation and migration in neutrophils. Glycoconj. J. 2008, 25, 357–374. [Google Scholar] [CrossRef] [PubMed]
- Park, J.W.; Park, W.J.; Kuperman, Y.; Boura-Halfon, S.; Pewzner-Jung, Y.; Futerman, A.H. Ablation of very long acyl chain sphingolipids causes hepatic insulin resistance in mice due to altered detergent-resistant membranes. Hepatology 2013, 57, 525–532. [Google Scholar] [CrossRef] [PubMed]
- Imgrund, S.; Hartmann, D.; Farwanah, H.; Eckhardt, M.; Sandhoff, R.; Degen, J.; Gieselmann, V.; Sandhoff, K.; Willecke, K. Adult ceramide synthase 2 (CERS2)-deficient mice exhibit myelin sheath defects, cerebellar degeneration, and hepatocarcinomas. J. Biol. Chem. 2009, 284, 33549–33560. [Google Scholar] [CrossRef] [PubMed]
- Stiban, J.; Perera, M. Very long chain ceramides interfere with C16-ceramide-induced channel formation: A plausible mechanism for regulating the initiation of intrinsic apoptosis. Biochim. Biophys. Acta 2015, 1848, 561–567. [Google Scholar] [CrossRef] [PubMed]
- Koivusalo, M.; Jansen, M.; Somerharju, P.; Ikonen, E. Endocytic trafficking of sphingomyelin depends on its acyl chain length. Mol. Biol. Cell 2007, 18, 5113–5123. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.S.; Rouser, G. The fatty acid composition of brain sphingolipids: Sphingomyelin, ceramide, cerebroside, and cerebroside sulfate. J. Lipid Res. 1964, 5, 339–342. [Google Scholar] [PubMed]
- Shaner, R.L.; Allegood, J.C.; Park, H.; Wang, E.; Kelly, S.; Haynes, C.A.; Sullards, M.C.; Merrill, A.H., Jr. Quantitative analysis of sphingolipids for lipidomics using triple quadrupole and quadrupole linear ion trap mass spectrometers. J. Lipid Res. 2009, 50, 1692–1707. [Google Scholar] [CrossRef] [PubMed]
- Loizides-Mangold, U.; David, F.P.; Nesatyy, V.J.; Kinoshita, T.; Riezman, H. Glycosylphosphatidylinositol anchors regulate glycosphingolipid levels. J. Lipid Res. 2012, 53, 1522–1534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christian, M.; Cermak, T.; Doyle, E.L.; Schmidt, C.; Zhang, F.; Hummel, A.; Bogdanove, A.J.; Voytas, D.F. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 2010, 186, 757–761. [Google Scholar] [CrossRef] [PubMed]
- Yamaji, T.; Hanada, K. Establishment of HeLa cell mutants deficient in sphingolipid-related genes using TALENs. PLoS ONE 2014, 9, e88124. [Google Scholar] [CrossRef] [PubMed]
- Mesika, A.; Ben-Dor, S.; Laviad, E.L.; Futerman, A.H. A new functional motif in Hox domain-containing ceramide synthases: Identification of a novel region flanking the Hox and TLC domains essential for activity. J. Biol. Chem. 2007, 282, 27366–27373. [Google Scholar] [CrossRef] [PubMed]
- Ohno, Y.; Suto, S.; Yamanaka, M.; Mizutani, Y.; Mitsutake, S.; Igarashi, Y.; Sassa, T.; Kihara, A. ELOVL1 production of C24 acyl-CoAs is linked to C24 sphingolipid synthesis. Proc. Natl. Acad. Sci. USA 2010, 107, 18439–18444. [Google Scholar] [CrossRef] [PubMed]
- Pewzner-Jung, Y.; Park, H.; Laviad, E.L.; Silva, L.C.; Lahiri, S.; Stiban, J.; Erez-Roman, R.; Brügger, B.; Sachsenheimer, T.; Wieland, F.; et al. A critical role for ceramide synthase 2 in liver homeostasis: I. alterations in lipid metabolic pathways. J. Biol. Chem. 2010, 285, 10902–10910. [Google Scholar] [CrossRef] [PubMed]
- Spassieva, S.D.; Mullen, T.D.; Townsend, D.M.; Obeid, L.M. Disruption of ceramide synthesis by CerS2 down-regulation leads to autophagy and the unfolded protein response. Biochem. J. 2009, 424, 273–283. [Google Scholar] [CrossRef] [PubMed]
- Fukasawa, M.; Nishijima, M.; Hanada, K. Genetic evidence for ATP-dependent endoplasmic reticulum-to-Golgi apparatus trafficking of ceramide for sphingomyelin synthesis in Chinese hamster ovary cells. J. Cell Biol. 1999, 144, 673–685. [Google Scholar] [CrossRef] [PubMed]
- Lippincott-Schwartz, J.; Yuan, L.C.; Bonifacino, J.S.; Klausner, R.D. Rapid redistribution of Golgi proteins into the ER in cells treated with brefeldin A: Evidence for membrane cycling from Golgi to ER. Cell 1989, 56, 801–813. [Google Scholar] [CrossRef]
- Casanova, J.E. Regulation of Arf activation: The Sec7 family of guanine nucleotide exchange factors. Traffic 2007, 8, 1476–1485. [Google Scholar] [CrossRef] [PubMed]
- Mullen, T.D.; Spassieva, S.; Jenkins, R.W.; Kitatani, K.; Bielawski, J.; Hannun, Y.A.; Obeid, L.M. Selective knockdown of ceramide synthases reveals complex interregulation of sphingolipid metabolism. J. Lipid Res. 2011, 52, 68–77. [Google Scholar] [CrossRef] [PubMed]
- Mesicek, J.; Lee, H.; Feldman, T.; Jiang, X.; Skobeleva, A.; Berdyshev, E.V.; Haimovitz-Friedman, A.; Fuks, Z.; Kolesnick, R. Ceramide synthases 2, 5, and 6 confer distinct roles in radiation-induced apoptosis in HeLa cells. Cell Signal. 2010, 22, 1300–1307. [Google Scholar] [CrossRef] [PubMed]
- Laviad, E.L.; Kelly, S.; Merrill, A.H., Jr.; Futerman, A.H. Modulation of ceramide synthase activity via dimerization. J. Biol. Chem. 2012, 287, 21025–21033. [Google Scholar] [CrossRef] [PubMed]
- Kumagai, K.; Yasuda, S.; Okemoto, K.; Nishijima, M.; Kobayashi, S.; Hanada, K. CERT mediates intermembrane transfer of various molecular species of ceramides. J. Biol. Chem. 2005, 280, 6488–6495. [Google Scholar] [CrossRef] [PubMed]
- Kok, J.W.; Babia, T.; Klappe, K.; Egea, G.; Hoekstra, D. Ceramide transport from endoplasmic reticulum to Golgi apparatus is not vesicle-mediated. Biochem. J. 1998, 333, 779–786. [Google Scholar] [CrossRef] [PubMed]
- Yamaji, T.; Nishikawa, K.; Hanada, K. Transmembrane BAX inhibitor motif containing (TMBIM) family proteins perturbs a trans-Golgi network enzyme, Gb3 synthase, and reduces Gb3 biosynthesis. J. Biol. Chem. 2010, 285, 35505–35518. [Google Scholar] [CrossRef] [PubMed]
- Cermak, T.; Doyle, E.L.; Christian, M.; Wang, L.; Zhang, Y.; Schmidt, C.; Baller, J.A.; Somia, N.V.; Bogdanove, A.J.; Voytas, D.F. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011, 39, e82. [Google Scholar] [CrossRef] [PubMed]
- Doyle, E.L.; Booher, N.J.; Standage, D.S.; Voytas, D.F.; Brendel, V.P.; Vandyk, J.K.; Bogdanove, A.J. TAL Effector-Nucleotide Targeter (TALE-NT) 2.0: Tools for TAL effector design and target prediction. Nucleic Acids Res. 2012, 40, 117–122. [Google Scholar] [CrossRef] [PubMed]
- Morita, S.; Kojima, T.; Kitamura, T. Plat-E: An efficient and stable system for transient packaging of retroviruses. Gene Ther. 2000, 7, 1063–1066. [Google Scholar] [CrossRef] [PubMed]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef] [PubMed]
- Hanada, K.; Hara, T.; Nishijima, M.; Kuge, O.; Dickson, R.C.; Nagiec, M.M. A mammalian homolog of the yeast LCB1 encodes a component of serine palmitoyltransferase, the enzyme catalyzing the first step in sphingolipid synthesis. J. Biol. Chem. 1997, 272, 32108–32114. [Google Scholar] [CrossRef] [PubMed]






© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yamaji, T.; Horie, A.; Tachida, Y.; Sakuma, C.; Suzuki, Y.; Kushi, Y.; Hanada, K. Role of Intracellular Lipid Logistics in the Preferential Usage of Very Long Chain-Ceramides in Glucosylceramide. Int. J. Mol. Sci. 2016, 17, 1761. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17101761
Yamaji T, Horie A, Tachida Y, Sakuma C, Suzuki Y, Kushi Y, Hanada K. Role of Intracellular Lipid Logistics in the Preferential Usage of Very Long Chain-Ceramides in Glucosylceramide. International Journal of Molecular Sciences. 2016; 17(10):1761. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17101761
Chicago/Turabian StyleYamaji, Toshiyuki, Aya Horie, Yuriko Tachida, Chisato Sakuma, Yusuke Suzuki, Yasunori Kushi, and Kentaro Hanada. 2016. "Role of Intracellular Lipid Logistics in the Preferential Usage of Very Long Chain-Ceramides in Glucosylceramide" International Journal of Molecular Sciences 17, no. 10: 1761. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17101761