Metabolomic Profiling of Plasma from Melioidosis Patients Using UHPLC-QTOF MS Reveals Novel Biomarkers for Diagnosis
Abstract
:1. Introduction
2. Results
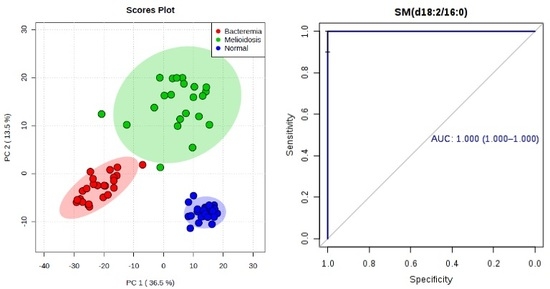
2.1. Metabolomic Profiling of Plasma Samples from Melioidosis Patients, Patients with Other Bacteremia and Controls
2.2. Omics-Based Statistical and Bioinformatic Analysis for Identification of Biomarkers
2.3. Diagnostic Performance of Metabolites
3. Discussion
4. Materials and Methods
4.1. Patient and Control Samples
4.2. Chemicals and Reagents
4.3. Sample Preparation
4.4. Untargeted Metabolomics Profiling of Patient Plasma Using UHPLC-ESI-QTOFMS
4.5. Data Processing and Statistical Analysis
4.6. Metabolite Identification
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| ACS | American Chemical Society |
| ANOVA | analysis of variance |
| ATII | alveolar type II pneumocytes |
| AUC | area-under-receiver operating characteristic curve |
| CE | collision energy |
| DPCC | dipalmitoylphosphatidylcholine |
| FA | fatty acid |
| FC | fold-change |
| IL-6 | interleukin-6 |
| IL-8 | interleukin-8 |
| LysoPE | lysophosphatidylethanolamine |
| MCCV | Monte Carlo Cross Validation |
| PC | phosphatidylcholine |
| PCA | principal component analysis |
| PIS | product ion scanning |
| PLA2 | phospholipase A2 |
| PlcH | phospholipase C |
| ROC | receiver operating characteristic curve |
| RT | retention time |
| SM | sphingomyelins |
| SIRS | systemic inflammatory response syndrome |
| SVM | support vector machines |
| TCA | tricarboxylic acid |
| TNF-α | Tumor necrosis factor-α |
| UHPLC-ESI-QTOFMS | ultra-high-performance liquid chromatography-electrospray ionization-quadruple time-of-flight-mass spectrometry |
| VIP | variable importance in the projection |
References
- Anuradha, K.; Meena, A.K.; Lakshmi, V. Isolation of Burkholderia pseudomallei from a case of septicaemia—A case report. Indian J. Med. Microbiol. 2003, 21, 129–132. [Google Scholar] [PubMed]
- Saravu, K.; Mukhopadhyay, C.; Vishwanath, S.; Valsalan, R.; Docherla, M.; Vandana, K.E.; Shastry, B.A.; Bairy, I.; Rao, S.P. Melioidosis in southern India: Epidemiological and clinical profile. Southeast. Asian J. Trop. Med. Public Health 2010, 41, 401–409. [Google Scholar] [PubMed]
- Issack, M.I.; Bundhun, C.D.; Gokhool, H. Melioidosis in Mauritius. Emerg. Infect. Dis. 2005, 11, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Inglis, T.J.; Rolim, D.B.; Sousa Ade, Q. Melioidosis in the Americas. Am. J. Trop Med. Hyg. 2006, 75, 947–954. [Google Scholar] [PubMed]
- Rolim, D.B.; Vilar, D.C.; Sousa, A.Q.; Miralles, I.S.; de Oliveira, D.C.; Harnett, G.; O’Reilly, L.; Howard, K.; Sampson, I.; Inglis, T.J. Melioidosis, northeastern Brazil. Emerg. Infect. Dis 2005, 11, 1458–1460. [Google Scholar] [CrossRef] [PubMed]
- Stewart, T.; Engelthaler, D.M.; Blaney, D.D.; Tuanyok, A.; Wangsness, E.; Smith, T.L.; Pearson, T.; Komatsu, K.K.; Keim, P.; Currie, B.J.; et al. Epidemiology and investigation of melioidosis, southern Arizona. Emerg. Infect. Dis. 2011, 17, 1286–1288. [Google Scholar] [CrossRef] [PubMed]
- Salam, A.P.; Khan, N.; Malnick, H.; Kenna, D.T.; Dance, D.A.; Klein, J.L. Melioidosis acquired by traveler to Nigeria. Emerg. Infect. Dis 2011, 17, 1296–1298. [Google Scholar] [CrossRef] [PubMed]
- Cuadros, J.; Gil, H.; Miguel, J.D.; Marabe, G.; Gomez-Herruz, T.A.; Lobo, B.; Marcos, R.; Anda, P. Case report: Melioidosis imported from west Africa to Europe. Am. J. Trop. Med. Hyg. 2011, 85, 282–284. [Google Scholar] [CrossRef] [PubMed]
- Currie, B.J.; Mayo, M.; Anstey, N.M.; Donohoe, P.; Haase, A.; Kemp, D.J. A cluster of melioidosis cases from an endemic region is clonal and is linked to the water supply using molecular typing of Burkholderia pseudomallei isolates. Am. J. Trop. Med. Hyg. 2001, 65, 177–179. [Google Scholar] [PubMed]
- Lau, S.K.; Chan, S.Y.; Curreem, S.O.; Hui, S.W.; Lau, C.C.; Lee, P.; Ho, C.C.; Martelli, P.; Woo, P.C. Burkholderia pseudomallei in soil samples from an oceanarium in Hong Kong detected using a sensitive PCR assay. Emerg. Microbes Infect. 2014, 3, e69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mays, E.E.; Ricketts, E.A. Melioidosis: Recrudescence associated with bronchogenic carcinoma twenty-six years following initial geographic exposure. Chest 1975, 68, 261–263. [Google Scholar] [CrossRef] [PubMed]
- Currie, B.J.; Fisher, D.A.; Howard, D.M.; Burrow, J.N.; Lo, D.; Selva-Nayagam, S.; Anstey, N.M.; Huffam, S.E.; Snelling, P.L.; Marks, P.J.; et al. Endemic melioidosis in tropical northern Australia: A 10-year prospective study and review of the literature. Clin. Infect. Dis. 2000, 31, 981–986. [Google Scholar] [CrossRef] [PubMed]
- Dance, D.A. Melioidosis. Curr. Opin. Infect. Dis. 2002, 15, 127–132. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.; Lau, S.K.; Woo, G.K.; Fung, A.M.; Ngan, A.H.; Hui, W.T.; Yuen, K.Y. Seronegative bacteremic melioidosis caused by Burkholderia pseudomallei with ambiguous biochemical profile: Clinical importance of accurate identification by 16s rRNA gene and groEL gene sequencing. J. Clin. Microbiol. 2003, 41, 3973–3977. [Google Scholar] [CrossRef] [PubMed]
- Cheng, A.C.; Currie, B.J. Melioidosis: Epidemiology, pathophysiology, and management. Clin. Microbiol. Rev. 2005, 18, 383–416. [Google Scholar] [CrossRef] [PubMed]
- Weissert, C.; Dollenmaier, G.; Rafeiner, P.; Riehm, J.; Schultze, D. Burkholderia pseudomallei misidentified by automated system. Emerg. Infect. Dis. 2009, 15, 1799–1801. [Google Scholar] [CrossRef] [PubMed]
- Godoy, D.; Randle, G.; Simpson, A.J.; Aanensen, D.M.; Pitt, T.L.; Kinoshita, R.; Spratt, B.G. Multilocus sequence typing and evolutionary relationships among the causative agents of melioidosis and glanders, Burkholderia pseudomallei and Burkholderia mallei. J. Clin. Microbiol. 2003, 41, 2068–2079. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.K.; Tang, B.S.; Curreem, S.O.; Chan, T.M.; Martelli, P.; Tse, C.W.; Wu, A.K.; Yuen, K.Y.; Woo, P.C. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for rapid identification of Burkholderia pseudomallei: Importance of expanding databases with pathogens endemic to different localities. J. Clin. Microbiol. 2012, 50, 3142–3143. [Google Scholar] [CrossRef] [PubMed]
- Inglis, T.J.; Healy, P.E.; Fremlin, L.J.; Golledge, C.L. Use of matrix-assisted laser desorption/ionization time-of-flight mass spectrometry analysis for rapid confirmation of Burkholderia pseudomallei in septicemic melioidosis. Am. J. Trop Med. Hyg. 2012, 86, 1039–1042. [Google Scholar] [CrossRef] [PubMed]
- Payne, G.W.; Vandamme, P.; Morgan, S.H.; Lipuma, J.J.; Coenye, T.; Weightman, A.J.; Jones, T.H.; Mahenthiralingam, E. Development of a recA gene-based identification approach for the entire Burkholderia genus. Appl. Environ. Microbiol. 2005, 71, 3917–3927. [Google Scholar] [CrossRef] [PubMed]
- Schmoock, G.; Ehricht, R.; Melzer, F.; Rassbach, A.; Scholz, H.C.; Neubauer, H.; Sachse, K.; Mota, R.A.; Saqib, M.; Elschner, M. DNA microarray-based detection and identification of Burkholderia mallei, Burkholderia pseudomallei and Burkholderia spp. Mol. Cell. Probes 2009, 23, 178–187. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.; Woo, G.K.; Lau, S.K.; Wong, S.S.; Yuen, K. Single gene target bacterial identification. groEL gene sequencing for discriminating clinical isolates of Burkholderia pseudomallei and Burkholderia thailandensis. Diagn. Microbiol. Infect. Dis. 2002, 44, 143–149. [Google Scholar] [CrossRef]
- Ho, C.C.; Lau, C.C.; Martelli, P.; Chan, S.Y.; Tse, C.W.; Wu, A.K.; Yuen, K.Y.; Lau, S.K.; Woo, P.C. Novel pan-genomic analysis approach in target selection for multiplex PCR identification and detection of Burkholderia pseudomallei, Burkholderia thailandensis, and Burkholderia cepacia complex species: A proof-of-concept study. J. Clin. Microbiol. 2011, 49, 814–821. [Google Scholar] [CrossRef] [PubMed]
- Puthucheary, S.D.; Anuar, A.S.; Tee, T.S. Burkholderia thailandensis whole cell antigen cross-reacts with B. pseudomallei antibodies from patients with melioidosis in an immunofluorescent assay. Southeast. Asian J. Trop. Med. Public Health 2010, 41, 395–400. [Google Scholar] [PubMed]
- Limmathurotsakul, D.; Chantratita, N.; Teerawattanasook, N.; Piriyagitpaiboon, K.; Thanwisai, A.; Wuthiekanun, V.; Day, N.P.; Cooper, B.; Peacock, S.J. Enzyme-linked immunosorbent assay for the diagnosis of melioidosis: Better than we thought. Clin. Infect. Dis. 2011, 52, 1024–1028. [Google Scholar] [CrossRef] [PubMed]
- Olivier, I.; Loots, D.T. A metabolomics approach to characterise and identify various Mycobacterium species. J. Microbiol. Methods 2012, 88, 419–426. [Google Scholar] [CrossRef] [PubMed]
- Olivier, I.; Loots, D.T. An overview of tuberculosis treatments and diagnostics. What role could metabolomics play? J. Cell. Tissue Res. 2011, 11, 2655–2671. [Google Scholar]
- Meissner-Roloff, R.J.; Koekemoer, G.; Warren, R.M.; Loots, D.T. A metabolomics investigation of a hyper- and hypo-virulent phenotype of Beijing lineage m. Tuberculosis. Metabolomics 2012, 8, 1194–1203. [Google Scholar] [CrossRef]
- Tam, E.W.; Chen, J.H.; Lau, E.C.; Ngan, A.H.; Fung, K.S.; Lee, K.C.; Lam, C.W.; Yuen, K.Y.; Lau, S.K.; Woo, P.C. Misidentification of Aspergillus nomius and Aspergillus tamarii as Aspergillus flavus: Characterization by internal transcribed spacer, β-tubulin, and calmodulin gene sequencing, metabolic fingerprinting, and matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 2014, 52, 1153–1160. [Google Scholar] [PubMed]
- To, K.K.; Fung, A.M.; Teng, J.L.; Curreem, S.O.; Lee, K.C.; Yuen, K.Y.; Lam, C.W.; Lau, S.K.; Woo, P.C. Characterization of a Tsukamurella pseudo-outbreak by phenotypic tests, 16s rRNA sequencing, pulsed-field gel electrophoresis, and metabolic footprinting. J. Clin. Microbiol. 2013, 51, 334–338. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.; Lam, C.W.; Tam, E.W.; Leung, C.K.; Wong, S.S.; Lau, S.K.; Yuen, K.Y. First discovery of two polyketide synthase genes for mitorubrinic acid and mitorubrinol yellow pigment biosynthesis and implications in virulence of Penicillium marneffei. PLoS Negl. Trop. Dis. 2012, 6, e1871. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.K.; Lam, C.W.; Curreem, S.O.; Lee, K.C.; Chow, W.N.; Lau, C.C.; Sridhar, S.; Wong, S.C.; Martelli, P.; Hui, S.W.; et al. Metabolomic profiling of Burkholderia pseudomallei using uhplc-esi-q-tof-ms reveals specific biomarkers including 4-methyl-5-thiazoleethanol and unique thiamine degradation pathway. Cell. Biosci. 2015, 5, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, K.C.; Tam, E.W.; Lo, K.C.; Tsang, A.K.; Lau, C.C.; To, K.K.; Chan, J.F.; Lam, C.W.; Yuen, K.Y.; Lau, S.K.; et al. Metabolomics analysis reveals specific novel tetrapeptide and potential anti-inflammatory metabolites in pathogenic Aspergillus species. Int. J. Mol. Sci. 2015, 16, 13850–13867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lau, S.K.; Lam, C.W.; Curreem, S.O.; Lee, K.C.; Lau, C.C.; Chow, W.N.; Ngan, A.H.; To, K.K.; Chan, J.F.; Hung, I.F.; et al. Identification of specific metabolites in culture supernatant of Mycobacterium tuberculosis using metabolomics: Exploration of potential biomarkers. Emerg. Microbes Infect. 2015, 4, e6. [Google Scholar] [CrossRef] [PubMed]
- Madigan, C.A.; Cheng, T.Y.; Layre, E.; Young, D.C.; McConnell, M.J.; Debono, C.A.; Murry, J.P.; Wei, J.R.; Barry, C.E., 3rd; Rodriguez, G.M.; et al. Lipidomic discovery of deoxysiderophores reveals a revised mycobactin biosynthesis pathway in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 2012, 109, 1257–1262. [Google Scholar] [CrossRef] [PubMed]
- Layre, E.; Lee, H.J.; Young, D.C.; Martinot, A.J.; Buter, J.; Minnaard, A.J.; Annand, J.W.; Fortune, S.M.; Snider, B.B.; Matsunaga, I.; et al. Molecular profiling of Mycobacterium tuberculosis identifies tuberculosinyl nucleoside products of the virulence-associated enzyme rv3378c. Proc. Natl. Acad. Sci. USA 2014, 111, 2978–2983. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.K.; Lee, K.C.; Curreem, S.O.; Chow, W.N.; To, K.K.; Hung, I.F.; Ho, D.T.; Sridhar, S.; Li, I.W.S.; Wu, A.L.; et al. Plasma metabolome changes in patients with tuberculosis using untargeted uhplc-qtof ms method reveal novel biomarkers for diagnosis. J. Clin. Microbiol. 2015, 53, 3750–3759. [Google Scholar] [PubMed]
- To, K.K.; Lee, K.C.; Wong, S.S.; Lo, K.C.; Lui, Y.M.; Jahan, A.S.; Wu, A.L.; Ke, Y.H.; Law, C.Y.; Sze, K.H.; et al. Lipid mediators of inflammation as novel plasma biomarkers to identify patients with bacteremia. J. Infect. 2015, 70, 433–444. [Google Scholar] [CrossRef] [PubMed]
- XCMS. Available online: http://www.bioconductor.org/packages/2.8/bioc/html/xcms.html (accessed on 30 November 2015).
- MetaboAnalyst 3.0—A Comprehersive Tool Suite Dor Metabolomic Data Analysis. Available online: http://www.metaboanalyst.ca/ (accessed on 1 December 2015).
- Indiveri, C.; Iacobazzi, V.; Tonazzi, A.; Giangregorio, N.; Infantino, V.; Convertini, P.; Console, L.; Palmieri, F. The mitochondrial carnitine/acylcarnitine carrier: Function, structure and physiopathology. Mol. Asp. Med. 2011, 32, 223–233. [Google Scholar] [CrossRef] [PubMed]
- Chin, C.Y.; Monack, D.M.; Nathan, S. Genome wide transcriptome profiling of a murine acute melioidosis model reveals new insights into how Burkholderia pseudomallei overcomes host innate immunity. BMC Genom. 2010, 11, 672. [Google Scholar] [CrossRef] [PubMed]
- Schmerler, D.; Neugebauer, S.; Ludewig, K.; Bremer-Streck, S.; Brunkhorst, F.M.; Kiehntopf, M. Targeted metabolomics for discrimination of systemic inflammatory disorders in critically ill patients. J. Lipid Res. 2012, 53, 1369–1375. [Google Scholar] [CrossRef] [PubMed]
- Tepperman, B.L.; Soper, B.D. The role of phospholipase A2 in calcium-ionophore-mediated injury to rat gastric mucosal cells. Dig. Dis. Sci. 1999, 44, 494–502. [Google Scholar] [CrossRef] [PubMed]
- Facciotti, F.; Ramanjaneyulu, G.S.; Lepore, M.; Sansano, S.; Cavallari, M.; Kistowska, M.; Forss-Petter, S.; Ni, G.; Colone, A.; Singhal, A.; et al. Peroxisome-derived lipids are self antigens that stimulate invariant natural killer T cells in the thymus. Nat. Immunol. 2012, 13, 474–480. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, N.; Ngwenya, B.Z. Activation of mouse peritoneal macrophages by lysophospholipids and ether derivatives of neutral lipids and phospholipids. Cancer Res. 1987, 47, 2008–2013. [Google Scholar] [PubMed]
- Endo, S.; Inada, K.; Nakae, H.; Takakuwa, T.; Yamada, Y.; Suzuki, T.; Taniguchi, S.; Yoshida, M.; Ogawa, M.; Teraoka, H. Plasma levels of type II phospholipase A2 and cytokines in patients with sepsis. Res. Commun. Mol. Pathol. Pharmacol. 1995, 90, 413–421. [Google Scholar] [PubMed]
- Gonzalez, E.; van Liempd, S.; Conde-Vancells, J.; Gutierrez-de Juan, V.; Perez-Cormenzana, M.; Mayo, R.; Berisa, A.; Alonso, C.; Marquez, C.A.; Barr, J.; et al. Serum uplc-ms/ms metabolic profiling in an experimental model for acute-liver injury reveals potential biomarkers for hepatotoxicity. Metabolomics 2012, 8, 997–1011. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Wang, H.; Xu, W.; Hao, D.; Du, L.; Zhao, X.; Sun, C. Metabolomic analysis of rat plasma following chronic low-dose exposure to dichlorvos. Hum. Exp. Toxicol. 2013, 32, 196–205. [Google Scholar] [CrossRef] [PubMed]
- Luberto, C.; Stonehouse, M.J.; Collins, E.A.; Marchesini, N.; El-Bawab, S.; Vasil, A.I.; Vasil, M.L.; Hannun, Y.A. Purification, characterization, and identification of a sphingomyelin synthase from Pseudomonas aeruginosa. PlcH is a multifunctional enzyme. J. Biol. Chem. 2003, 278, 32733–32743. [Google Scholar] [CrossRef] [PubMed]
- Korbsrisate, S.; Tomaras, A.P.; Damnin, S.; Ckumdee, J.; Srinon, V.; Lengwehasatit, I.; Vasil, M.L.; Suparak, S. Characterization of two distinct phospholipase c enzymes from Burkholderia pseudomallei. Microbiology 2007, 153, 1907–1915. [Google Scholar] [CrossRef] [PubMed]
- Tuanyok, A.; Tom, M.; Dunbar, J.; Woods, D.E. Genome-wide expression analysis of Burkholderia pseudomallei infection in a hamster model of acute melioidosis. Infect. Immun. 2006, 74, 5465–5476. [Google Scholar] [CrossRef] [PubMed]
- Theodorou, I.G.; Botelho, D.; Schwander, S.; Zhang, J.; Chung, K.F.; Tetley, T.D.; Shaffer, M.S.; Gow, A.; Ryan, M.P.; Porter, A.E. Static and dynamic microscopy of the chemical stability and aggregation state of silver nanowires in components of murine pulmonary surfactant. Environ. Sci. Technol. 2015, 49, 8048–8056. [Google Scholar] [CrossRef] [PubMed]
- Creuwels, L.A.; van Golde, L.M.; Haagsman, H.P. The pulmonary surfactant system: Biochemical and clinical aspects. Lung 1997, 175, 1–39. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.; Popov, V.; Patel, J.; Eaves-Pyles, T. Burkholderia mallei and Burkholderia pseudomallei stimulate differential inflammatory responses from human alveolar type II cells (ATII) and macrophages. Front. Cell. Infect. Microbiol. 2012, 2, 165. [Google Scholar] [CrossRef] [PubMed]
- The R Project for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 30 November 2015).
- Basanta, M.; Jarvis, R.M.; Xu, Y.; Blackburn, G.; Tal-Singer, R.; Woodcock, A.; Singh, D.; Goodacre, R.; Thomas, C.L.; Fowler, S.J. Non-invasive metabolomic analysis of breath using differential mobility spectrometry in patients with chronic obstructive pulmonary disease and healthy smokers. Analyst 2010, 135, 315–320. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Broadhurst, D.I.; Wilson, M.; Wishart, D.S. Translational biomarker discovery in clinical metabolomics: an introductory tutorial. Metabolomics 2013, 9, 280–299. [Google Scholar] [CrossRef] [PubMed]
- Scripps Center for Meatbolomics – METLIN. Available online: http://metlin.scripps.edu/ (accessed on 1 December 2015).
- The Human Metabolome Database. Available online: http://www.hmdb.ca/ (accessed on 1 December 2015).
- Massbank. Available online: http://www.massbank.jp/ (accessed on 1 December 2015).
- LIPID Metabolites and Pathways Strategy (LIPID MAPS®) – Lipidomics Gateway. Available online: http://www.lipidmaps.org/ (accessed on 1 December 2015).
- KEGG: Kyoto Encyclopedia of Genes and Genomes. Available online: http://www.genome.jp/kegg/ (accessed on 1 December 2015).









| Compound | Experimental Mass, m/z | Ion | Retention Time (min) | MS/MS Fragment Masses | Elemental Composition | Metabolite Class |
|---|---|---|---|---|---|---|
| l-Hexanoylcarnitine | 260.1842 | [M + H]+ | 4.47 | 60.0808, 85.0285, 99.0803, 144.1018, 201.1117 | C13H25NO4 | acylcarnitine |
| l-Octanoylcarnitine | 288.2157 | [M + H]+ | 6.78 | 60.0805, 85.0283, 127.1110, 144.1019, 229.1438 | C15H29NO4 | acylcarnitine |
| 2-Decenoylcarnitine | 314.2326 | [M + H]+ | 7.84 | 60.0806, 85.0281, 144.1015, 153.1257, 255.1591 | C17H31NO4 | acylcarnitine |
| Decanoylcarnitine | 316.2476 | [M + H]+ | 8.62 | 60.0806, 85.0284, 144.1017, 155.1424, 257.1748 | C17H33NO4 | acylcarnitine |
| Trans-2-dodecenoylcarnitine | 342.2636 | [M + H]+ | 9.38 | 60.0807, 85.0283, 144.1019, 181.1584, 283.1880 | C19H35NO4 | acylcarnitine |
| Dodecanoylcarnitine | 344.2775 | [M + H]+ | 10.20 | 60.0806, 85.0284, 144.1008, 183.1735, 285.2088 | C19H37NO4 | acylcarnitine |
| LysoPE(16:0/0:0) | 454.2934 | [M + H]+ | 14.64 | 44.0496, 62.0598, 216.0642, 239.2361, 257.2530, 313.2740, 393.2423, 436.2774 | C21H44NO7P | lysophosphatidylethanolamine |
| LysoPE(0:0/18:0) | 482.3244 | [M + H]+ | 16.94 | 44.0494, 216.0628, 267.2644, 285.2747, 341.3060 | C23H48NO7P | lysophosphatidylethanolamine |
| LysoPE(18:0/0:0) | 482.3251 | [M + H]+ | 17.61 | 44.0497, 62.0600, 216.0618, 267.2672, 285.2777, 341.3058, 421.2718, 464.3112 | C23H48NO7P | lysophosphatidylethanolamine |
| SM(d16:1/16:0) | 675.5444 | [M + H]+ | 27.88 | 60.0808, 104.1072, 184.0735, 236.2355 | C37H75N2O6P | sphingomyelins |
| SM(d18:2/16:0) | 701.5605 | [M + H]+ | 28.64 | 60.0802, 104.1068, 184.0736, 262.2575, 683.5484 | C39H77N2O6P | sphingomyelins |
| PC(16:0/16:0) | 734.5616 | [M + H]+ | 30.58 | 60.0801, 104.1060, 184.0727, 478.3251, 496.3353 | C40H80NO8P | phosphatidylcholine |
| Significant Metabolites | Melioidosis vs. Bacteremia | Melioidosis vs. Control without Active Infections | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AUC a | 95% CI b | Sensitivity (%) | Specificity (%) | p-Value c | Fold-Change | AUC a | 95% CI b | Sensitivity (%) | Specificity (%) | p-Value c | Fold-Change | |
| l-Hexanoylcarnitine | 0.665 | 0.497–0.805 | 81.8 | 50.0 | 1.12 × 10−2 | 2.32 ↑d | 0.849 | 0.728–0.959 | 83.3 | 81.8 | 1.10 × 10−3 | 3.33 ↑d |
| l-Octanoylcarnitine | 0.856 | 0.710–0.964 | 86.4 | 79.2 | 6.34 × 10−4 | 3.49 ↑ | 0.827 | 0.693–0.942 | 83.3 | 86.4 | 2.90 × 10−3 | 2.76 ↑ |
| 2-Decenoylcarnitine | 0.839 | 0.700–0.937 | 77.3 | 79.2 | 4.77 × 10−3 | 3.63 ↑ | 0.829 | 0.682–0.952 | 90.0 | 81.8 | 1.90 × 10−3 | 3.56 ↑ |
| Decanoylcarnitine | 0.850 | 0.728–0.972 | 77.3 | 87.5 | 1.46 × 10−4 | 3.53 ↑ | 0.821 | 0.680–0.949 | 83.3 | 77.3 | 1.45 × 10−3 | 2.66 ↑ |
| Trans-2-dodecenoylcarnitine | 0.850 | 0.736–0.965 | 77.3 | 79.2 | 1.20 × 10−5 | 2.40 ↑ | 0.886 | 0.776–0.965 | 90.0 | 72.7 | 5.43 × 10−8 | 2.88 ↑ |
| Dodecanoylcarnitine | 0.822 | 0.700–0.935 | 72.7 | 91.7 | 3.10 × 10−5 | 2.46 ↑ | 0.741 | 0.583–0.877 | 76.7 | 72.7 | 9.60 × 10−3 | 1.66 ↑ |
| LysoPE(16:0/0:0) | 0.979 | 0.947–1.000 | 90.9 | 95.8 | 1.51 × 10−10 | 5.20 ↑ | 0.812 | 0.672–0.933 | 73.3 | 81.8 | 2.56 × 10−6 | 2.01 ↑ |
| LysoPE(0:0/18:0) | 0.994 | 0.982–1.000 | 95.5 | 100.0 | 6.08 × 10−10 | 7.51 ↑ | 0.819 | 0.658–0.926 | 90.0 | 77.3 | 1.96 × 10−6 | 2.23 ↑ |
| LysoPE(18:0/0:0) | 0.998 | 0.993–1.000 | 100.0 | 95.8 | 1.77 × 10−11 | 6.09 ↑ | 0.856 | 0.725–0.958 | 90.0 | 77.3 | 1.41 × 10−7 | 2.16 ↑ |
| SM(d16:1/16:0) | 0.968 | 0.927–1.000 | 90.9 | 91.7 | 7.19 × 10−10 | 3.41 ↑ | 0.884 | 0.783–0.983 | 83.3 | 77.3 | 2.67 × 10−8 | 2.16 ↑ |
| SM(d18:2/16:0) | 0.998 | 0.993–1.000 | 100.0 | 91.7 | 1.88 × 10−12 | 3.32 ↑ | 1.000 | 1.000–1.000 | 96.7 | 100.0 | 1.28 × 10−13 | 2.65 ↑ |
| PC(16:0/16:0) | 0.835 | 0.695–0.976 | 77.3 | 87.5 | 7.01 × 10−5 | 9.64 ↑ | 0.870 | 0.724–0.989 | 93.3 | 81.8 | 2.77 × 10−6 | 21.72 ↑ |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lau, S.K.P.; Lee, K.-C.; Lo, G.C.S.; Ding, V.S.Y.; Chow, W.-N.; Ke, T.Y.H.; Curreem, S.O.T.; To, K.K.W.; Ho, D.T.Y.; Sridhar, S.; et al. Metabolomic Profiling of Plasma from Melioidosis Patients Using UHPLC-QTOF MS Reveals Novel Biomarkers for Diagnosis. Int. J. Mol. Sci. 2016, 17, 307. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17030307
Lau SKP, Lee K-C, Lo GCS, Ding VSY, Chow W-N, Ke TYH, Curreem SOT, To KKW, Ho DTY, Sridhar S, et al. Metabolomic Profiling of Plasma from Melioidosis Patients Using UHPLC-QTOF MS Reveals Novel Biomarkers for Diagnosis. International Journal of Molecular Sciences. 2016; 17(3):307. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17030307
Chicago/Turabian StyleLau, Susanna K. P., Kim-Chung Lee, George C. S. Lo, Vanessa S. Y. Ding, Wang-Ngai Chow, Tony Y. H. Ke, Shirly O. T. Curreem, Kelvin K. W. To, Deborah T. Y. Ho, Siddharth Sridhar, and et al. 2016. "Metabolomic Profiling of Plasma from Melioidosis Patients Using UHPLC-QTOF MS Reveals Novel Biomarkers for Diagnosis" International Journal of Molecular Sciences 17, no. 3: 307. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17030307