Natural Killer Cells—An Epigenetic Perspective of Development and Regulation
Abstract
:1. Natural Killer Cells
2. Epigenetic Modifications
3. Epigenetic Modifications in NK Cell Development
4. Epigenetic Regulation of NK Effector Function
5. Epigenetic Regulation of NK Cell Receptors
5.1. Killer Cell Immunoglobin-Like Receptors (KIR)
5.2. NKG2D
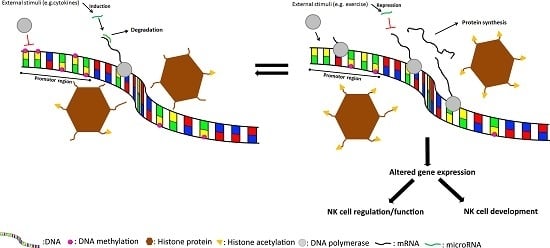
6. External Stimuli
7. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Caligiuri, M.A. Human natural killer cells. Blood 2008, 112, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Cooper, M.A.; Fehniger, T.A.; Turner, S.C.; Chen, K.S.; Ghaheri, B.A.; Ghayur, T.; Carson, W.E.; Caligiuri, M.A. Human natural killer cells: A unique innate immunoregulatory role for the CD56(bright) subset. Blood 2001, 97, 3146–3151. [Google Scholar] [CrossRef] [PubMed]
- Chan, H.-W.; Kurago, Z.B.; Stewart, C.A.; Wilson, M.J.; Martin, M.P.; Mace, B.E.; Carrington, M.; Trowsdale, J.; Lutz, C.T. DNA methylation maintains allele-specific KIR gene expression in human natural killer cells. J. Exp. Med. 2003, 197, 245–255. [Google Scholar] [CrossRef] [PubMed]
- Espinoza, J.L.; Takami, A.; Yoshioka, K.; Nakata, K.; Sato, T.; Kasahara, Y.; Nakao, S. Human microRNA-1245 down-regulates the NKG2D receptor in natural killer cells and impairs NKG2D-mediated functions. Haematologica 2012, 97, 1295–1303. [Google Scholar] [CrossRef] [PubMed]
- Schroder, K.; Hertzog, P.J.; Ravasi, T.; Hume, D.A. Interferon-gamma: an overview of signals, mechanisms and functions. J. Leukoc. Biol. 2004, 75, 163–189. [Google Scholar] [CrossRef] [PubMed]
- Subleski, J.; Weiss, J.M.; Wiltrout, R.H.; Ortaldo, J.R. NK and NKT cells: The innate-adaptive interface including humoral responses: NK cytokines and chemokines. In Natural Killer Cells: Basic Science and Clinical Application; Lotze, M.T., Thomson, A.W., Eds.; Elsevier/Academic Press: London, UK, 2010; p. 258. [Google Scholar]
- Walzer, T.; Jaeger, S.; Chaix, J.; Vivier, E. Natural killer cells: From CD3−NKp46+ to post-genomics meta-analyses. Curr. Opin. Immunol. 2007, 19, 365–372. [Google Scholar] [CrossRef] [PubMed]
- Luetke-Eversloh, M.; Hammer, Q.; Durek, P.; Nordström, K.; Gasparoni, G.; Pink, M.; Hamann, A.; Walter, J.; Chang, H.-D.; Dong, J.; et al. Human cytomegalovirus drives epigenetic imprinting of the IFNG locus in NKG2Chi natural killer cells. PLoS Pathog. 2014, 10, e1004441. [Google Scholar] [CrossRef] [PubMed]
- Gleeson, M.; Bosch, J. The human immune system: The role of natural killer cells in innate immunity. In Exercise Immunology; Gleeson, M., Bishop, N., Walsh, N., Eds.; Taylor & Francis Group: London, UK, 2013; pp. 34–35. [Google Scholar]
- Stern-Ginossar, N.; Mandelboim, O. Receptors on NK cells. In Natural Killer Cells: Basic Science and Clinical Application; Lotze, M.T., Thomson, A.W., Eds.; Elsevier/Academic Press: London, UK, 2010; pp. 155–165. [Google Scholar]
- Armas, L.R.; de Podack, E.R. Natural killer cytolytic activity: Lytic granule components. In Natural Killer Cells: Basic Science and Clinical Application; Lotze, M.T., Thomson, A.W., Eds.; Elsevier/Academic Press: London, UK, 2010; pp. 217–220. [Google Scholar]
- Trapani, J.A.; Sutton, V.R. Granzyme B: Pro-apoptotic, antiviral and antitumor functions. Curr. Opin. Immunol. 2003, 15, 533–543. [Google Scholar] [CrossRef]
- Holliday, R. Epigenetics: A historical overview. Epigenetics 2006, 1, 76–80. [Google Scholar] [CrossRef] [PubMed]
- Allis, C.D.; Jenuwein, T.; Reinberg, D. Overview and concepts. In Epigenetics; Allis, C.D., Jenuwein, T., Reinberg, D., Eds.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2007; pp. 25–56. [Google Scholar]
- Watt, F.; Molloy, P.L. Cytosine methylation prevents binding to DNA of a HeLa cell transcription factor required for optimal expression of the adenovirus major late promoter. Genes Dev. 1988, 2, 1136–1143. [Google Scholar] [CrossRef] [PubMed]
- Boyes, J.; Bird, A. DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell 1991, 64, 1123–1134. [Google Scholar] [CrossRef]
- Costello, J.F.; Futscher, B.W.; Tano, K.; Graunke, D.M.; Pieper, R.O. Graded methylation in the promoter and body of the O6-methylguanine DNA methyltransferase (MGMT) gene correlates with MGMT expression in human glioma cells. J. Biol. Chem. 1994, 269, 17228–17237. [Google Scholar] [PubMed]
- Dupont, C.; Armant, D.R.; Brenner, C.A. Epigenetics: Definition, mechanisms and clinical perspective. Semin. Reprod. Med. 2009, 27, 351–357. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Sánchez, A.; Baragaño Raneros, A.; Carvajal Palao, R.; Sanz, A.B.; Ortiz, A.; Ortega, F.; Suárez-Álvarez, B.; López-Larrea, C. DNA demethylation and histone H3K9 acetylation determine the active transcription of the NKG2D gene in human CD8+ T and NK cells. Epigenetics 2013, 8, 66–78. [Google Scholar] [CrossRef] [PubMed]
- Santourlidis, S.; Graffmann, N.; Christ, J.; Uhrberg, M. Lineage-specific transition of histone signatures in the killer cell Ig-like receptor locus from hematopoietic progenitor to NK cells. J. Immunol. 2008, 180, 418–425. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.-N.; Lin, J.; Wang, L.-L.; Yu, L. Demethylating treatment suppresses natural killer cell cytolytic activity. Mol. Immunol. 2009, 46, 2064–2070. [Google Scholar] [CrossRef] [PubMed]
- Cichocki, F.; Felices, M.; McCullar, V.; Presnell, S.R.; Al-Attar, A.; Lutz, C.T.; Miller, J.S. Cutting edge: MicroRNA-181 promotes human NK cell development by regulating Notch signaling. J. Immunol. 2011, 187, 6171–6175. [Google Scholar] [CrossRef] [PubMed]
- Haraguchi, K.; Suzuki, T.; Koyama, N.; Kumano, K.; Nakahara, F.; Matsumoto, A.; Yokoyama, Y.; Sakata-Yanagimoto, M.; Masuda, S.; Takahashi, T.; et al. Notch activation induces the generation of functional NK cells from human cord blood CD34− positive cells devoid of IL-15. J. Immunol. 2009, 182, 6168–6178. [Google Scholar] [CrossRef] [PubMed]
- Felices, M.; Ankarlo, D.E.M.; Lenvik, T.R.; Nelson, H.H.; Blazar, B.R.; Verneris, M.R.; Miller, J.S. Notch signaling at later stages of NK cell development enhances KIR expression and functional maturation. J. Immunol. 2014, 193, 3344–3354. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.C.; Beilke, J.N.; Lanier, L.L. Adaptive immune features of natural killer cells. Nature 2009, 457, 557–561. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Zhang, T.; Hwang, I.; Kim, A.; Nitschke, L.; Kim, M.; Scott, J.M.; Kamimura, Y.; Lanier, L.L.; Kim, S. Epigenetic modification and antibody-dependent expansion of memory-like NK cells in human cytomegalovirus-infected individuals. Immunity 2015, 42, 431–442. [Google Scholar] [CrossRef] [PubMed]
- Schlums, H.; Cichocki, F.; Tesi, B.; Theorell, J.; Beziat, V.; Holmes, T.D.; Han, H.; Chiang, S.C.C.; Foley, B.; Mattsson, K.; et al. Cytomegalovirus infection drives adaptive epigenetic diversification of NK cells with altered signaling and effector function. Immunity 2015, 42, 443–456. [Google Scholar] [CrossRef] [PubMed]
- Ogbomo, H.; Michaelis, M.; Kreuter, J.; Doerr, H.W.; Cinatl, J. Histone deacetylase inhibitors suppress natural killer cell cytolytic activity. FEBS Lett. 2007, 581, 1317–1322. [Google Scholar] [CrossRef] [PubMed]
- Schmiedel, B.J.; Arélin, V.; Gruenebach, F.; Krusch, M.; Schmidt, S.M.; Salih, H.R. Azacytidine impairs NK cell reactivity while decitabine augments NK cell responsiveness toward stimulation. Int. J. Cancer 2011, 128, 2911–2922. [Google Scholar] [CrossRef] [PubMed]
- Cooley, S.; Miller, J.S. Clinical trials of NK cells for cancer: Killer immunoglobulin-like receptors. In Natural Killer Cells: Basic Science and Clinical Application; Lotze, M.T., Thomson, A.W., Eds.; Elsevier/Academic Press: London, UK, 2010; p. 557. [Google Scholar]
- Santourlidis, S.; Trompeter, H.-I.; Weinhold, S.; Eisermann, B.; Meyer, K.L.; Wernet, P.; Uhrberg, M. Crucial role of DNA methylation in determination of clonally distributed killer cell Ig-like receptor expression patterns in NK cells. J. Immunol. 2002, 169, 4253–4261. [Google Scholar] [CrossRef] [PubMed]
- Cichocki, F.; Lenvik, T.; Sharma, N.; Yun, G.; Anderson, S.K.; Miller, J.S. Cutting edge: KIR antisense transcripts are processed into a 28-base PIWI-like RNA in human NK cells. J. Immunol. 2010, 185, 2009–2012. [Google Scholar] [CrossRef] [PubMed]
- Davies, G.E.; Locke, S.M.; Wright, P.W.; Li, H.; Hanson, R.J.; Miller, J.S.; Anderson, S.K. Identification of bidirectional promoters in the human KIR genes. Genes Immun. 2007, 8, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Jahn, J.; Spielau, M.; Brandsch, C.; Stangl, G.I.; Delank, K.-S.; Bähr, I.; Berreis, T.; Wrann, C.D.; Kielstein, H. Decreased NK cell functions in obesity can be reactivated by fat mass reduction. Obesity 2015, 23, 2233–2241. [Google Scholar] [CrossRef] [PubMed]
- Gleeson, M. Effects of exercise on innate immune function: Natural killer cell cytotoxic acitivity. In Exercise Immunology; Gleeson, M., Bishop, N., Walsh, N., Eds.; Taylor & Francis Group: London, UK, 2013; pp. 116–120. [Google Scholar]
- Hupkes, M.; Jonsson, M.K.B.; Scheenen, W.J.; van Rotterdam, W.; Sotoca, A.M.; van Someren, E.P.; van der Heyden, M.A.G.; van Veen, T.A.; van Ravestein-van Os, R.I.; Bauerschmidt, S.; et al. Epigenetics: DNA demethylation promotes skeletal myotube maturation. FASEB J. 2011, 25, 3861–3872. [Google Scholar] [CrossRef] [PubMed]
- Radom-Aizik, S.; Zaldivar, F.; Leu, S.-Y.; Adams, G.R.; Oliver, S.; Cooper, D.M. Effects of exercise on microRNA expression in young males peripheral blood mononuclear cells. Clin. Transl. Sci. 2012, 5, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Alexander, M.R.; Owens, G.K. Epigenetic control of smooth muscle cell differentiation and phenotypic switching in vascular development and disease. Annu. Rev. Physiol. 2012, 74, 13–40. [Google Scholar] [CrossRef] [PubMed]
- Rönn, T.; Volkov, P.; Davegårdh, C.; Dayeh, T.; Hall, E.; Olsson, A.H.; Nilsson, E.; Tornberg, A.; Dekker Nitert, M.; Eriksson, K.-F.; et al. A six months exercise intervention influences the genome-wide DNA methylation pattern in human adipose tissue. PLoS Genet. 2013, 9, e1003572. [Google Scholar] [CrossRef] [PubMed]
- Abel, J.L.; Rissman, E.F. Running-induced epigenetic and gene expression changes in the adolescent brain. Int. J. Dev. Neurosci. 2013, 31, 382–390. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, K.; Takeoka, M.; Mori, M.; Hashimoto, S.; Sakurai, A.; Nose, H.; Higuchi, K.; Itano, N.; Shiohara, M.; Oh, T.; et al. Exercise effects on methylation of ASC gene. Int. J. Sports Med. 2010, 31, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Zimmer, P.; Baumann, F.T.; Bloch, W.; Schenk, A.; Koliamitra, C.; Jensen, P.; Mierau, A.; Hülsdünker, T.; Reinart, N.; Hallek, M.; et al. Impact of exercise on pro inflammatory cytokine levels and epigenetic modulations of tumor-competitive lymphocytes in Non-Hodgkin-Lymphoma patients-randomized controlled trial. Eur. J. Haematol. 2014, 93, 527–532. [Google Scholar] [CrossRef] [PubMed]
- Zimmer, P.; Bloch, W.; Schenk, A.; Zopf, E.M.; Hildebrandt, U.; Streckmann, F.; Beulertz, J.; Koliamitra, C.; Schollmayer, F.; Baumann, F. Exercise-induced Natural Killer Cell Activation is Driven by Epigenetic Modifications. Int. J. Sports Med. 2015, 36, 510–515. [Google Scholar] [CrossRef] [PubMed]
- Sapolsky, R.M.; Romero, L.M.; Munck, A.U. How do glucocorticoids influence stress responses? Integrating permissive, suppressive, stimulatory, and preparative actions. Endocr. Rev. 2000, 21, 55–89. [Google Scholar] [CrossRef] [PubMed]
- Schoneveld, O.; Cidlowski, J.A. Glucocorticoids and Immunity: Mechanisms of Regulation. In Psychoneuroimmunology, 4th ed.; Ader, R., Ed.; Elsevier/Academic Press: Boston, UK, 2007; pp. 45–61. [Google Scholar]
- Krukowski, K.; Eddy, J.; Kosik, K.L.; Konley, T.; Janusek, L.W.; Mathews, H.L. Glucocorticoid dysregulation of natural killer cell function through epigenetic modification. Brain Behav. Immun. 2011, 25, 239–249. [Google Scholar] [CrossRef] [PubMed]
- Bush, K.A.; Krukowski, K.; Eddy, J.L.; Janusek, L.W.; Mathews, H.L. Glucocorticoid receptor mediated suppression of natural killer cell activity: Identification of associated deacetylase and corepressor molecules. Cell Immunol. 2012, 275, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Eddy, J.L.; Krukowski, K.; Janusek, L.; Mathews, H.L. Glucocorticoids regulate natural killer cell function epigenetically. Cell Immunol. 2014, 290, 120–130. [Google Scholar] [CrossRef] [PubMed]

© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schenk, A.; Bloch, W.; Zimmer, P. Natural Killer Cells—An Epigenetic Perspective of Development and Regulation. Int. J. Mol. Sci. 2016, 17, 326. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17030326
Schenk A, Bloch W, Zimmer P. Natural Killer Cells—An Epigenetic Perspective of Development and Regulation. International Journal of Molecular Sciences. 2016; 17(3):326. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17030326
Chicago/Turabian StyleSchenk, Alexander, Wilhelm Bloch, and Philipp Zimmer. 2016. "Natural Killer Cells—An Epigenetic Perspective of Development and Regulation" International Journal of Molecular Sciences 17, no. 3: 326. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17030326