EIYMNVPV Motif is Essential for A1CF Nucleus Localization and A1CF (-8aa) Promotes Proliferation of MDA-MB-231 Cells via Up-Regulation of IL-6
Abstract
:1. Introduction
2. Results
2.1. Bioinformatics Analysis of A1CF
2.2. The Deletion of the EIYMNVPV Motif by Alternative Splicing is Conserved Across Species
2.3. EIYMNVPV Motif is Essential for A1CF Nucleus Localization
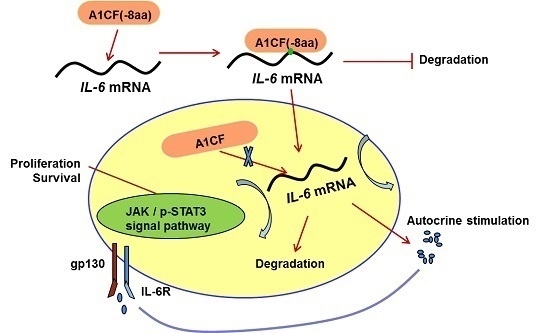
2.4. A1CF (-8aa) Promotes Proliferation of MDA-MB-231 Cells
2.5. IL-6 Was Involved in A1CF (-8aa)-Promoted Proliferation of MDA-MB-231 Cells
3. Discussion
4. Materials and Methods
4.1. Plasmid Construction
4.2. Cell Culture and Transfection
- siRNA1:sense5′-GCAGACCCAAGAUACCCUA-3′, antisense 3′-UAGGGUAUCUUGGGUCUGC-5′;
- siRNA2: sense5′-GGUCUGGCAUGUGACCAUU-3′, antisense3′-AAUGGUCACAUGCCAGACC-5′;
- siRNA3:sense5′-GGGUGAUAGUGACAGUGAA-3′, antisense3′-UUCACUGUCACUAUCACCC-5′.
4.3. RNA Extraction, RT-PCR, Quantitative RT-PCR (qPCR)
- has-A1CF (-8aa)/A1CF (F: GCCAAGTTTATGATCCCACC; R: CAGTCCTCT CACTCCCGCA);
- hsa-IL-6 (F: GCCACTCACCTCTTCAGAACG; R: CAGTGCCTCTTTGCTG CTTTC).
4.4. Protein Extraction and Western Blot Analysis
4.5. Immunofluorescence
4.6. 5-Ethynyl-20-Deoxyuridine (EdU) Assays
4.7. Statistical Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| dsRBM | Double strand RNA binding motif |
| PCNA | Proliferating cell nuclear antigen |
| A1CF | Apobec-1 complementation factor |
| cDNA | Complementary DNA |
| qPCR | Quantitative reverse-transcription polymerase chain reaction |
| RRM | RNA recognition motifs |
| apoB | Apolipoprotein-B |
| EdU | 5-Ethynyl-20-Deoxyuridine |
| IL-6 | Interleukin-6 |
References
- Lellek, H.; Kirsten, R.; Diehl, I.; Apostel, F.; Buck, F.; Greeve, J. Purification and molecular cloning of a novel essential component of the apolipoprotein B mRNA editing enzyme-complex. J. Boil. Chem. 2000, 275, 19848–19856. [Google Scholar] [CrossRef] [PubMed]
- Maris, C.; Masse, J.; Chester, A.; Navaratnam, N.; Allain, F.H. NMR structure of the apoB mRNA stem-loop and its interaction with the C to U sediting APOBEC1 complementary factor. RNA 2005, 11, 173–186. [Google Scholar] [CrossRef] [PubMed]
- Mehta, A.; Kinter, M.T.; Sherman, N.E.; Driscoll, D.M. Molecular cloning of apobec-1 complementation factor, a novel RNA-binding protein involved in the editing of apolipoprotein B mRNA. Mol. Cell. Biol. 2000, 20, 1846–1854. [Google Scholar] [CrossRef] [PubMed]
- Teng, B.; Burant, C.F.; Davidson, N.O. Molecular cloning of an apolipoprotein B messenger RNA editing protein. Science 1993, 260, 1816–1819. [Google Scholar] [CrossRef] [PubMed]
- Backus, J.W.; Smith, H.C. Apolipoprotein B mRNA sequences 3’ of the editing site are necessary and sufficient for editing and editosome assembly. Nucleic Acids Res. 1991, 19, 6781–6786. [Google Scholar] [CrossRef] [PubMed]
- Chester, A.; Scott, J.; Anant, S.; Navaratnam, N. RNA editing: Cytidine to uridine conversion in apolipoprotein B mRNA. Biochim. Biophys. Acta 2000, 1494, 1–13. [Google Scholar] [CrossRef]
- Shah, R.R.; Knott, T.J.; Legros, J.E.; Navaratnam, N.; Greeve, J.C.; Scott, J. Sequence requirements for the editing of apolipoprotein B mRNA. J. Boil. Chem. 1991, 266, 16301–16304. [Google Scholar]
- Smith, H.C.; Sowden, M.P. Base-modification mRNA editing through deamination—The good, the bad and the unregulated. Trends Genet. TIG 1996, 12, 418–424. [Google Scholar] [CrossRef]
- Paschoud, S.; Dogar, A.M.; Kuntz, C.; Grisoni-Neupert, B.; Richman, L.; Kuhn, L.C. Destabilization of interleukin-6 mRNA requires a putative RNA stem-loop structure, an AU-rich element, and the RNA-binding protein AUF1. Mol. Cell. Biol. 2006, 26, 8228–8241. [Google Scholar] [CrossRef] [PubMed]
- Blanc, V.; Henderson, J.O.; Newberry, E.P.; Kennedy, S.; Luo, J.; Davidson, N.O. Targeted deletion of the murine apobec-1 complementation factor (acf) gene results in embryonic lethality. Mol. Cell. Biol. 2005, 25, 7260–7269. [Google Scholar] [CrossRef] [PubMed]
- Blanc, V.; Sessa, K.J.; Kennedy, S.; Luo, J.; Davidson, N.O. Apobec-1 complementation factor modulates liver regeneration by post-transcriptional regulation of interleukin-6 mRNA stability. J. Boil. Chem. 2010, 285, 19184–19192. [Google Scholar] [CrossRef] [PubMed]
- Mehta, A.; Driscoll, D.M. Identification of domains in apobec-1 complementation factor required for RNA binding and apolipoprotein-B mRNA editing. RNA 2002, 8, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Dance, G.S.; Sowden, M.P.; Cartegni, L.; Cooper, E.; Krainer, A.R.; Smith, H.C. Two proteins essential for apolipoprotein B mRNA editing are expressed from a single gene through alternative splicing. J. Boil. Chem. 2002, 277, 12703–12709. [Google Scholar] [CrossRef] [PubMed]
- Fossat, N.; Tourle, K.; Radziewic, T.; Barratt, K.; Liebhold, D.; Studdert, J.B.; Power, M.; Jones, V.; Loebel, D.A.; Tam, P.P. C to U RNA editing mediated by APOBEC1 requires RNA-binding protein RBM47. EMBO Rep. 2014, 15, 903–910. [Google Scholar] [CrossRef] [PubMed]
- Vanharanta, S.; Marney, C.B.; Shu, W.; Valiente, M.; Zou, Y.; Mele, A.; Darnell, R.B.; Massague, J. Loss of the multifunctional RNA-binding protein RBM47 as a source of selectable metastatic traits in breast cancer. eLife 2014, 3. [Google Scholar] [CrossRef] [PubMed]
- Knupfer, H.; Preiss, R. Significance of interleukin-6 (IL-6) in breast cancer (review). Breast Cancer Res. Treat. 2007, 102, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Ara, T.; Declerck, Y.A. Interleukin-6 in bone metastasis and cancer progression. Eur. J. Cancer 2010, 46, 1223–1231. [Google Scholar] [CrossRef] [PubMed]
- Chang, Q.; Daly, L.; Bromberg, J. The IL-6 feed-forward loop: A driver of tumorigenesis. Semin. Immunol. 2014, 26, 48–53. [Google Scholar] [CrossRef] [PubMed]
- Chomarat, P.; Banchereau, J.; Davoust, J.; Palucka, A.K. IL-6 switches the differentiation of monocytes from dendritic cells to macrophages. Nat. Immunol. 2000, 1, 510–514. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Xu, F.; Lu, T.; Duan, Z.; Zhang, Z. Interleukin-6 signaling pathway in targeted therapy for cancer. Cancer Treat. Rev. 2012, 38, 904–910. [Google Scholar] [CrossRef] [PubMed]
- Itou, J.; Tanaka, S.; Sato, F.; Akiyama, R.; Kawakami, Y.; Toi, M. An optical labeling-based proliferation assay system reveals the paracrine effect of interleukin-6 in breast cancer. Biochim. Biophys. Acta 2015, 1853, 27–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yadav, A.; Kumar, B.; Datta, J.; Teknos, T.N.; Kumar, P. IL-6 promotes head and neck tumor metastasis by inducing epithelial-mesenchymal transition via the JAK-STAT3-SNAIL signaling pathway. Mol. Cancer Res. MCR 2011, 9, 1658–1667. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.P.; Li, J.; Tewari, A.K. Inflammation and prostate cancer: The role of interleukin 6 (IL-6). BJU Int. 2014, 113, 986–992. [Google Scholar] [CrossRef] [PubMed]
- Waldner, M.J.; Neurath, M.F. Master regulator of intestinal disease: IL-6 in chronic inflammation and cancer development. Semin. Immunol. 2014, 26, 75–79. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.W.; Sun, Y.M. The IL-6/JAK/STAT3 pathway: Potential therapeutic strategies in treating colorectal cancer (Review). Int. J. Oncol. 2014, 44, 1032–1040. [Google Scholar] [PubMed]
- Lin, S.; Gan, Z.; Han, K.; Yao, Y.; Min, D. Interleukin-6 as a prognostic marker for breast cancer: A meta-analysis. Tumori 2015, 101, 535–541. [Google Scholar] [CrossRef] [PubMed]
- Blanc, V.; Kennedy, S.; Davidson, N.O. A novel nuclear localization signal in the auxiliary domain of apobec-1 complementation factor regulates nucleocytoplasmic import and shuttling. J. Boil. Chem. 2003, 278, 41198–41204. [Google Scholar] [CrossRef] [PubMed]
- Chiang, C.P.; Lang, M.J.; Liu, B.Y.; Wang, J.T.; Leu, J.S.; Hahn, L.J.; Kuo, M.Y. Expression of proliferating cell nuclear antigen (PCNA) in oral submucous fibrosis, oral epithelial hyperkeratosis and oral epithelial dysplasia in Taiwan. Oral Oncol. 2000, 36, 353–359. [Google Scholar] [CrossRef]
- Srinivasan, M.; Jewell, S.D. Quantitative estimation of PCNA, c-myc, EGFR and TGF-α in oral submucous fibrosis—An immunohistochemical study. Oral Oncol. 2001, 37, 461–467. [Google Scholar] [CrossRef]
- Heo, T.H.; Wahler, J.; Suh, N. Potential therapeutic implications of IL-6/IL-6R/gp130-targeting agents in breast cancer. Oncotarget 2016. [Google Scholar] [CrossRef] [PubMed]
- Ayub, A.; Yip, W.K.; Seow, H.F. Dual treatments targeting IGF-1R, PI3K, mTORC or MEK synergize to inhibit cell growth, induce apoptosis, and arrest cell cycle at G1 phase in MDA-MB-231 cell line. Biomed. Pharmacother. 2015, 75, 40–50. [Google Scholar] [CrossRef] [PubMed]
- Nakahara, S.; Tsutsumi, K.; Zuinen, T.; Ohta, Y. FilGAP, a Rho-ROCK-regulated GAP for Rac, controls adherens junctions in MDCK cells. J. Cell Sci. 2015, 128, 2047–2056. [Google Scholar] [CrossRef] [PubMed]





© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, L.; Hao, J.; Yuan, Y.; Peng, R.; Wang, H.; Ni, D.; Gu, Y.; Huang, L.; Mao, Z.; Lyu, Z.; et al. EIYMNVPV Motif is Essential for A1CF Nucleus Localization and A1CF (-8aa) Promotes Proliferation of MDA-MB-231 Cells via Up-Regulation of IL-6. Int. J. Mol. Sci. 2016, 17, 811. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17060811
Zhou L, Hao J, Yuan Y, Peng R, Wang H, Ni D, Gu Y, Huang L, Mao Z, Lyu Z, et al. EIYMNVPV Motif is Essential for A1CF Nucleus Localization and A1CF (-8aa) Promotes Proliferation of MDA-MB-231 Cells via Up-Regulation of IL-6. International Journal of Molecular Sciences. 2016; 17(6):811. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17060811
Chicago/Turabian StyleZhou, Li, Jin Hao, Yue Yuan, Rui Peng, Honglian Wang, Dongsheng Ni, Yuping Gu, Liyuan Huang, Zhaomin Mao, Zhongshi Lyu, and et al. 2016. "EIYMNVPV Motif is Essential for A1CF Nucleus Localization and A1CF (-8aa) Promotes Proliferation of MDA-MB-231 Cells via Up-Regulation of IL-6" International Journal of Molecular Sciences 17, no. 6: 811. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17060811