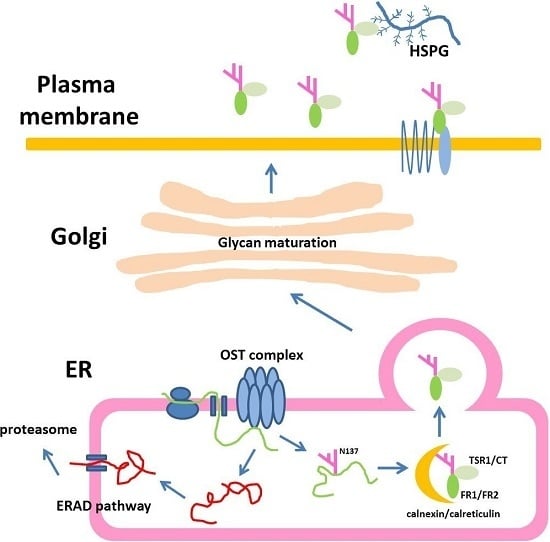
N-Glycosylation of Human R-Spondin 1 Is Required for Efficient Secretion and Stability but Not for Its Heparin Binding Ability
Abstract
:1. Introduction
2. Results
2.1. Human Rspo1 Is N-Glycosylated at Asn137 in Vivo
2.2. Rspo1 N-Glycosylation at Asn137 Affects Its Secretion
2.3. N-Glycosylation of Human Rspo3 Also Affects Its Secretion in Vivo
2.4. Introduction of the Rspo1 N-Glycosylation Site Enhances the Accumulation of Human Rspo2 in Media and Rescues the Accumulation Defect of Nonglycosylated Rspo2
2.5. Introduction of the Rspo2 N-Glycosylation Site Rescues the Accumulation Defects of Nonglycosylated Rspo1 and Rspo3 Mutants
2.6. Effect of N-Glycans on the Stability of Intracellular Rspo1
2.7. Effect of N-Glycan on the Stability of Secreted Rspos
2.8. Prevention of N-Glycosylation at N137 in Human Rspo1 and Rspo3 Did Not Affect Their Heparin-Binding Ability
3. Discussion
4. Experimental Section
4.1. Plasmid Construction and Site-Directed Mutagenesis
4.2. Cell Culture and Transfection
4.3. Cell Fractionation and Western Blot
4.4. Chemical Treatment and Deglycosylation
4.5. SuperTopFlash Reporter Assay
4.6. Pulse-Chase Experiments
4.7. Stability of Secreted Rspos
4.8. Heparin Binding Assay
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Kamata, T.; Katsube, K.; Michikawa, M.; Yamada, M.; Takada, S.; Mizusawa, H. R-spondin, a novel gene with thrombospondin type 1 domain, was expressed in the dorsal neural tube and affected in Wnts mutants. Biochim. Biophys. Acta 2004, 1676, 51–62. [Google Scholar] [CrossRef] [PubMed]
- Kazanskaya, O.; Glinka, A.; del Barco, B.I.; Stannek, P.; Niehrs, C.; Wu, W. R-spondin2 is a secreted activator of Wnt/beta-catenin signaling and is required for Xenopus myogenesis. Dev. Cell 2004, 7, 525–534. [Google Scholar] [CrossRef] [PubMed]
- Zebisch, M.; Xu, Y.; Krastev, C.; MacDonald, B.T.; Chen, M.; Gilbert, R.J.; He, X.; Jones, E.Y. Structural and molecular basis of ZNRF3/RNF43 transmembrane ubiquitin ligase inhibition by the Wnt agonist R-spondin. Nat. Commun. 2013, 4, 2787. [Google Scholar] [CrossRef] [PubMed]
- Nam, J.S.; Turcotte, T.J.; Smith, P.F.; Choi, S.; Yoon, J.K. Mouse cristin/R-spondin family proteins are novel ligands for the Frizzled 8 and LRP6 receptors and activate β-catenin-dependent gene expression. J. Biol. Chem. 2006, 281, 13247–13257. [Google Scholar] [CrossRef] [PubMed]
- Parma, P.; Radi, O.; Vidal, V.; Chaboissier, M.C.; Dellambra, E.; Valentini, S.; Guerra, L.; Schedl, A.; Camerino, G. R-spondin1 is essential in sex determination, skin differentiation and malignancy. Nat. Genet. 2006, 38, 1304–1309. [Google Scholar] [CrossRef] [PubMed]
- Blaydon, D.C.; Ishii, Y.; O’Toole, E.A.; Unsworth, H.C.; Teh, M.T.; Ruschendorf, F.; Sinclair, C.; Hopsu-Havu, V.K.; Tidman, N.; Moss, C.; et al. The gene encoding R-spondin 4 (Rspo4), a secreted protein implicated in Wnt signaling, is mutated in inherited anonychia. Nat. Genet. 2006, 38, 1245–1247. [Google Scholar] [CrossRef] [PubMed]
- Bergmann, C.; Senderek, J.; Anhuf, D.; Thiel, C.T.; Ekici, A.B.; Poblete-Gutierrez, P.; van Steensel, M.; Seelow, D.; Nurnberg, G.; Schild, H.H.; et al. Mutations in the gene encoding the Wnt-signaling component R-spondin 4 (Rspo4) cause autosomal recessive anonychia. Am. J. Hum. Genet. 2006, 79, 1105–1109. [Google Scholar] [CrossRef] [PubMed]
- Bell, S.M.; Schreiner, C.M.; Wert, S.E.; Mucenski, M.L.; Scott, W.J.; Whitsett, J.A. R-spondin 2 is required for normal laryngeal-tracheal, lung and limb morphogenesis. Development 2008, 135, 1049–1058. [Google Scholar] [CrossRef] [PubMed]
- Yamada, W.; Nagao, K.; Horikoshi, K.; Fujikura, A.; Ikeda, E.; Inagaki, Y.; Kakitani, M.; Tomizuka, K.; Miyazaki, H.; Suda, T.; et al. Craniofacial malformation in R-spondin2 knockout mice. Biochem. Biophys. Res. Commun. 2009, 381, 453–458. [Google Scholar] [CrossRef] [PubMed]
- Aoki, M.; Mieda, M.; Ikeda, T.; Hamada, Y.; Nakamura, H.; Okamoto, H. R-spondin3 is required for mouse placental development. Dev. Biol. 2007, 301, 218–226. [Google Scholar] [CrossRef] [PubMed]
- Kazanskaya, O.; Ohkawara, B.; Heroult, M.; Wu, W.; Maltry, N.; Augustin, H.G.; Niehrs, C. The Wnt signaling regulator R-spondin 3 promotes angioblast and vascular development. Development 2008, 135, 3655–3664. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.A.; Kakitani, M.; Zhao, J.; Oshima, T.; Tang, T.; Binnerts, M.; Liu, Y.; Boyle, B.; Park, E.; Emtage, P.; et al. Mitogenic influence of human R-spondin1 on the intestinal epithelium. Science 2005, 309, 1256–1259. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Vries, R.G.; Snippert, H.J.; van de Wetering, M.; Barker, N.; Stange, D.E.; van Es, J.H.; Abo, A.; Kujala, P.; Peters, P.J.; et al. Single LGR5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 2009, 459, 262–265. [Google Scholar] [CrossRef] [PubMed]
- Ootani, A.; Li, X.; Sangiorgi, E.; Ho, Q.T.; Ueno, H.; Toda, S.; Sugihara, H.; Fujimoto, K.; Weissman, I.L.; Capecchi, M.R.; et al. Sustained in vitro intestinal epithelial culture within a Wnt-dependent stem cell niche. Nat. Med. 2009, 15, 701–706. [Google Scholar] [CrossRef] [PubMed]
- Carmon, K.S.; Gong, X.; Lin, Q.; Thomas, A.; Liu, Q. R-spondins function as ligands of the orphan receptors LGR4 and LGR5 to regulate Wnt/β-catenin signaling. Proc. Natl. Acad. Sci. USA 2011, 108, 11452–11457. [Google Scholar] [CrossRef] [PubMed]
- De Lau, W.; Barker, N.; Low, T.Y.; Koo, B.K.; Li, V.S.; Teunissen, H.; Kujala, P.; Haegebarth, A.; Peters, P.J.; van de Wetering, M.; et al. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signaling. Nature 2011, 476, 293–297. [Google Scholar] [CrossRef] [PubMed]
- Glinka, A.; Dolde, C.; Kirsch, N.; Huang, Y.L.; Kazanskaya, O.; Ingelfinger, D.; Boutros, M.; Cruciat, C.M.; Niehrs, C. LGR4 and LGR5 are R-spondin receptors mediating Wnt/β-catenin and Wnt/PCP signaling. EMBO Rep. 2011, 12, 1055–1061. [Google Scholar] [CrossRef] [PubMed]
- Hao, H.X.; Xie, Y.; Zhang, Y.; Charlat, O.; Oster, E.; Avello, M.; Lei, H.; Mickanin, C.; Liu, D.; Ruffner, H.; et al. ZNRF3 promotes Wnt receptor turnover in an R-spondin-sensitive manner. Nature 2012, 485, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Koo, B.K.; Spit, M.; Jordens, I.; Low, T.Y.; Stange, D.E.; van de Wetering, M.; van Es, J.H.; Mohammed, S.; Heck, A.J.; Maurice, M.M.; et al. Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature 2012, 488, 665–669. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Xu, Y.; Rajashankar, K.R.; Robev, D.; Nikolov, D.B. Crystal structures of LGR4 and its complex with R-spondin1. Structure 2013, 21, 1683–1689. [Google Scholar] [CrossRef] [PubMed]
- Peng, W.C.; de Lau, W.; Madoori, P.K.; Forneris, F.; Granneman, J.C.; Clevers, H.; Gros, P. Structures of Wnt-antagonist ZNRF3 and its complex with R-spondin 1 and implications for signaling. PLoS ONE 2013, 8, e83110. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Huang, B.; Zhang, S.; Yu, X.; Wu, W.; Wang, X. Structural basis for R-spondin recognition by LGR4/5/6 receptors. Genes Dev. 2013, 27, 1339–1344. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.H.; Chen, X.; Lin, Z.; Fang, D.; He, X. The structural basis of R-spondin recognition by LGR5 and RNF43. Genes Dev. 2013, 27, 1345–1350. [Google Scholar] [CrossRef] [PubMed]
- Peng, W.C.; de Lau, W.; Forneris, F.; Granneman, J.C.; Huch, M.; Clevers, H.; Gros, P. Structure of stem cell growth factor R-spondin 1 in complex with the ectodomain of its receptor LGR5. Cell Rep. 2013, 3, 1885–1892. [Google Scholar] [CrossRef] [PubMed]
- Ohkawara, B.; Glinka, A.; Niehrs, C. Rspo3 binds syndecan 4 and induces Wnt/PCP signaling via clathrin-mediated endocytosis to promote morphogenesis. Dev. Cell 2011, 20, 303–314. [Google Scholar] [CrossRef] [PubMed]
- Moremen, K.W.; Tiemeyer, M.; Nairn, A.V. Vertebrate protein glycosylation: Diversity, synthesis and function. Nat. Rev. Mol. Cell Boil. 2012, 13, 448–462. [Google Scholar] [CrossRef] [PubMed]
- Imperiali, B.; Rickert, K.W. Conformational implications of asparagine-linked glycosylation. Proc. Natl. Acad. Sci. USA 1995, 92, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Bause, E. Structural requirements of N-glycosylation of proteins. Studies with proline peptides as conformational probes. Biochem. J. 1983, 209, 331–336. [Google Scholar] [CrossRef] [PubMed]
- Mellquist, J.L.; Kasturi, L.; Spitalnik, S.L.; Shakin-Eshleman, S.H. The amino acid following an asn-X-Ser/Thr sequon is an important determinant of N-linked core glycosylation efficiency. Biochemistry 1998, 37, 6833–6837. [Google Scholar] [CrossRef] [PubMed]
- Kelleher, D.J.; Gilmore, R. An evolving view of the eukaryotic oligosaccharyltransferase. Glycobiology 2006, 16, 47R–62R. [Google Scholar] [CrossRef] [PubMed]
- Helenius, A.; Aebi, M. Roles of N-linked glycans in the endoplasmic reticulum. Annu. Rev. Biochem. 2004, 73, 1019–1049. [Google Scholar] [CrossRef] [PubMed]
- Helenius, A.; Aebi, M. Intracellular functions of N-linked glycans. Science 2001, 291, 2364–2369. [Google Scholar] [CrossRef] [PubMed]
- Hoseki, J.; Ushioda, R.; Nagata, K. Mechanism and components of endoplasmic reticulum-associated degradation. J. Biochem. 2010, 147, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Van de Wetering, M.; Sancho, E.; Verweij, C.; de Lau, W.; Oving, I.; Hurlstone, A.; van der Horn, K.; Batlle, E.; Coudreuse, D.; Haramis, A.P.; et al. The β-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell 2002, 111, 241–250. [Google Scholar] [CrossRef]
- Veeman, M.T.; Slusarski, D.C.; Kaykas, A.; Louie, S.H.; Moon, R.T. Zebrafish prickle, a modulator of noncanonical Wnt/Fz signaling, regulates gastrulation movements. Curr. Biol. 2003, 13, 680–685. [Google Scholar] [CrossRef]
- Heifetz, A.; Keenan, R.W.; Elbein, A.D. Mechanism of action of tunicamycin on the UDP-GlcNAc:Dolichyl-phosphate Glc-NAc-1-phosphate transferase. Biochemistry 1979, 18, 2186–2192. [Google Scholar] [CrossRef] [PubMed]
- Saul, R.; Ghidoni, J.J.; Molyneux, R.J.; Elbein, A.D. Castanospermine inhibits alpha-glucosidase activities and alters glycogen distribution in animals. Proc. Natl. Acad. Sci. USA 1985, 82, 93–97. [Google Scholar] [CrossRef] [PubMed]
- Hammond, C.; Braakman, I.; Helenius, A. Role of N-linked oligosaccharide recognition, glucose trimming, and calnexin in glycoprotein folding and quality control. Proc. Natl. Acad. Sci. USA 1994, 91, 913–917. [Google Scholar] [CrossRef] [PubMed]
- Maley, F.; Trimble, R.B.; Tarentino, A.L.; Plummer, T.H., Jr. Characterization of glycoproteins and their associated oligosaccharides through the use of endoglycosidases. Anal. Biochem. 1989, 180, 195–204. [Google Scholar] [CrossRef]
- Smith, M.H.; Ploegh, H.L.; Weissman, J.S. Road to ruin: Targeting proteins for degradation in the endoplasmic reticulum. Science 2011, 334, 1086–1090. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dieterich, D.C.; Link, A.J.; Graumann, J.; Tirrell, D.A.; Schuman, E.M. Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT). Proc. Natl. Acad. Sci. USA 2006, 103, 9482–9487. [Google Scholar] [CrossRef] [PubMed]
- Mitra, N.; Sinha, S.; Ramya, T.N.; Surolia, A. N-linked oligosaccharides as outfitters for glycoprotein folding, form and function. Trends Biochem. Sci. 2006, 31, 156–163. [Google Scholar] [CrossRef] [PubMed]
- Hanson, S.R.; Culyba, E.K.; Hsu, T.L.; Wong, C.H.; Kelly, J.W.; Powers, E.T. The core trisaccharide of an N-linked glycoprotein intrinsically accelerates folding and enhances stability. Proc. Natl. Acad. Sci. USA 2009, 106, 3131–3136. [Google Scholar] [CrossRef] [PubMed]
- Moad, H.E.; Pioszak, A.A. Reconstitution of R-spondin:LGR4:ZNRF3 adult stem cell growth factor signaling complexes with recombinant proteins produced in Escherichia coli. Biochemistry 2013, 52, 7295–7304. [Google Scholar] [CrossRef] [PubMed]
- Baron, R.; Kneissel, M. Wnt signaling in bone homeostasis and disease: From human mutations to treatments. Nat. Med. 2013, 19, 179–192. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Brennan, T.A.; Russell, E.; Kim, J.H.; Egan, K.P.; Chen, Q.; Israelite, C.; Schultz, D.C.; Johnson, F.B.; Pignolo, R.J. R-spondin 1 promotes vibration-induced bone formation in mouse models of osteoporosis. J. Mol. Med. 2013, 91, 1421–1429. [Google Scholar] [CrossRef] [PubMed]
- Freeze, H.H. Genetic defects in the human glycome. Nat. Rev. Genet. 2006, 7, 537–551. [Google Scholar] [CrossRef] [PubMed]
- Park, C.; Zhang, J. Genome-wide evolutionary conservation of N-glycosylation sites. Mol. Biol. Evol. 2011, 28, 2351–2357. [Google Scholar] [CrossRef] [PubMed]
- Imperiali, B.; O’Connor, S.E. Effect of N-linked glycosylation on glycopeptide and glycoprotein structure. Curr. Opin. Chem. Biol. 1999, 3, 643–649. [Google Scholar] [CrossRef]
- Chang, C.F.; Tsai, J.N. Functional analysis of mucin type O-glycosylation for R-spondin 1. Int. J. Mol. Sci. 2016. in preparation. [Google Scholar]
- Tran, D.T.; Ten Hagen, K.G. Mucin-type O-glycosylation during development. J. Biol. Chem. 2013, 288, 6921–6929. [Google Scholar] [CrossRef] [PubMed]
- Hofsteenge, J.; Huwiler, K.G.; Macek, B.; Hess, D.; Lawler, J.; Mosher, D.F.; Peter-Katalinic, J. C-mannosylation and O-fucosylation of the thrombospondin type 1 module. J. Biol. Chem. 2001, 276, 6485–6498. [Google Scholar] [CrossRef] [PubMed]














© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, C.-F.; Hsu, L.-S.; Weng, C.-Y.; Chen, C.-K.; Wang, S.-Y.; Chou, Y.-H.; Liu, Y.-Y.; Yuan, Z.-X.; Huang, W.-Y.; Lin, H.; et al. N-Glycosylation of Human R-Spondin 1 Is Required for Efficient Secretion and Stability but Not for Its Heparin Binding Ability. Int. J. Mol. Sci. 2016, 17, 937. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17060937
Chang C-F, Hsu L-S, Weng C-Y, Chen C-K, Wang S-Y, Chou Y-H, Liu Y-Y, Yuan Z-X, Huang W-Y, Lin H, et al. N-Glycosylation of Human R-Spondin 1 Is Required for Efficient Secretion and Stability but Not for Its Heparin Binding Ability. International Journal of Molecular Sciences. 2016; 17(6):937. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17060937
Chicago/Turabian StyleChang, Chiung-Fang, Li-Sung Hsu, Chieh-Yu Weng, Chih-Kai Chen, Shu-Ying Wang, Yi-Hwa Chou, Yan-Yu Liu, Zi-Xiu Yuan, Wen-Ying Huang, Ho Lin, and et al. 2016. "N-Glycosylation of Human R-Spondin 1 Is Required for Efficient Secretion and Stability but Not for Its Heparin Binding Ability" International Journal of Molecular Sciences 17, no. 6: 937. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms17060937