Metabolic Features of Multiple Myeloma
Abstract
:1. Introduction
2. General Cancer Metabolism
3. Glucose Metabolism in multiple myeloma (MM)
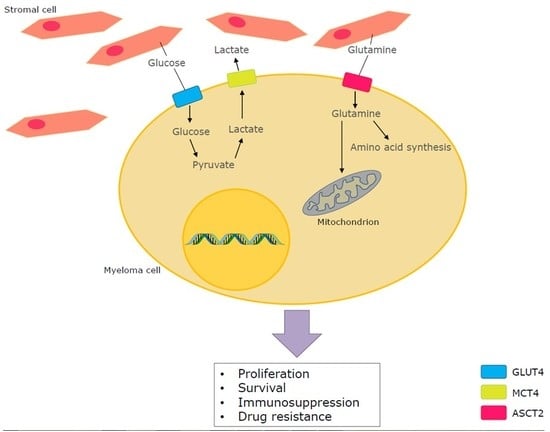
4. Glutamine Metabolism in MM
5. Drug Resistance in MM
5.1. Standard-of-Care Drugs
5.2. Immunotherapy
6. Conclusions and Future Perspectives
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| αKG | Alpha Ketoglutarate |
| ALL | Acute Lymphoblastic Leukaemia |
| 6-AN | 6-Aminonicotinamide |
| ASCT2 | Neutral Amino Acid Transporter 2 |
| ATP | Adenosine Triphosphate |
| BM | Bone Marrow |
| BMSC | Bone Marrow Stromal Cells |
| 3BP | 3-Bromopyruvate |
| CAM-DR | Cell Adhesion-Mediated Drug Resistance |
| CAR | Chimeric Antigen Receptor |
| CHC | α-Cyano-4-Hydroxycinnamic |
| DC | Dendritic Cell |
| DCA | Dichloroacetate |
| 2DG | 2-Deoxyglucose |
| DM | Dimethyl |
| ECM | Extracellular Matrix |
| EGFR | Epidermal Growth Factor |
| ERK1/2 | Extracellular signal-regulated Kinase |
| FAD | Flavin Adenine Dinucleotide |
| 18F-FDG | Fluorine-labelled Fluorodeoxyglucose |
| FGF-2 | Fibroblast Growth Factor-2 |
| FL | Fas Ligand |
| GLDH | Glutamate Dehydrogenase |
| GLS1 | Glutaminase 1 |
| GLS2 | Glutaminase 2 |
| GPNA | l-γ-Glutamyl-p-Nitroanilide |
| GS | Glutamine Synthetase |
| HKII | Hexokinase II |
| HMCL | Human Myeloma Cell Line |
| IAP | Inhibitors of Apoptosis Proteins |
| IGF-1 | Insulin-like Growth Factor-1 |
| IL-1β | Interleukin-1β |
| IL-6 | Interleukin-6 |
| IMiDs | Immunomodulatory |
| JAK | Janus Kinase |
| LAT1 | human L-type Amino acid Transporter 1 |
| LDH | Lactate Dehydrogenase |
| LDHA | Lactate Dehydrogenase A |
| MAPK | Mitogen-Activated Protein Kinase |
| MCT | Monocarboxylate Transporter |
| MGUS | Monoclonal Gammopathy of Undetermined Significance |
| MM | Multiple Myeloma |
| MSO | Methionine Sulfoximine |
| MYC | Myelocytomatosis oncogene cellular homolog |
| NAD | Nicotinamide Adenine Dinucleotide |
| NADH | Nicotinamide Adenine Dinucleotide Hydrogen |
| NH4+ | Ammonia |
| NK | Natural Killer |
| OAF | Osteoclast Activating Factors |
| OC | Osteoclast |
| OXPHOS | Oxidative Phosphorylation |
| PDH | Pyruvate Dehydrogenase |
| PEP | Phosphoenolpyruvate |
| PET | Positron Emission Tomography |
| PI3K | Phosphoinositide-3 Kinase |
| PK | Pyruvate Kinase |
| PPP | Pentose Phosphate Pathway |
| PRPP | 5-Phospho-α-Ribosyl-1-Pyrophosphate |
| RANKL | Receptor Activator of Nuclear factor κB Ligand |
| SNAT1 | Sodium-coupled Neutral Amino acid Transporter 1 |
| STAT3 | Signal Transducer and Activator of Transcription 3 |
| TCA | Tricarboxylic Acid cycle |
| TME | Tumour Micro-Environment |
| TNF | Tumour Necrosis Factor |
| TNF-β | Tumour Necrosis Factor-β |
| TRAIL | TNF-Related Apoptosis-Inducing Ligand |
| VCAM-1 | Vascular Cell Adhesion Molecule 1 |
| VDAC | Voltage-Dependent Anion Channel |
| VEGF | Vascular Endothelial Growth Factor |
References
- Kristinsson, S.Y.; Björkholm, M.; Goldin, L.R.; Blimark, C.; Mellqvist, U.H.; Wahlin, A.; Turesson, I.; Landgren, O. Patterns of hematologic malignancies and solid tumors among 37,838 first-degree relatives of 13,896 multiple myeloma patients in Sweden. Int. J. Cancer 2009, 125, 2147–2150. [Google Scholar] [CrossRef] [PubMed]
- Kawano, Y.; Moschetta, M.; Manier, S.; Glavey, S.; Görgün, GT.; Roccaro, A.M.; Anderson, K.C.; Ghobrial, I.M. Targeting the bone marrow microenvironment in multiple myeloma. Immunol. Rev. 2015, 263, 160–172. [Google Scholar] [CrossRef] [PubMed]
- Kühnel, A.; Blau, O.; Nogai, K.A.; Blau, I.W. The Warburg effect in Multiple Myeloma and its microenvironment. Med. Res. Arch. 2017, 5, 1–16. [Google Scholar]
- Rajkumar, S.V. Evolving diagnostic criteria for multiple myeloma. Hematol. Am. Soc. Hematol. Educ. Program 2015, 2015, 272–278. [Google Scholar] [CrossRef] [PubMed]
- Ramsenthaler, C.; Kane, P.; Gao, W.; Siegert, R.J.; Edmonds, P.M.; Schey, S.A.; Higginson, I.J. Prevalence of symptoms in patients with multiple myeloma: A systematic review and meta-analysis. Eur. J. Haematol. 2016, 97, 416–429. [Google Scholar] [CrossRef] [PubMed]
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.K.; Rajkumar, S.V.; Dispenzieri, A.; Lacy, M.Q.; Hayman, S.R.; Buadi, F.K.; Zeldenrust, S.R.; Dingli, D.; Russell, S.J.; Lust, J.A.; et al. Improved survival in multiple myeloma and the impact of novel therapies. Blood 2008, 111, 2516–2520. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, G.; Richardson, P.G.; Anderson, K.C. Promising therapies in multiple myeloma. Blood 2015, 126, 300–310. [Google Scholar] [CrossRef] [PubMed]
- Moreau, P.; Attal, M.; Facon, T. Frontline therapy of multiple myeloma. Blood 2015, 125, 3076–3084. [Google Scholar] [CrossRef] [PubMed]
- Russo, A.; Saide, A.; Smaldone, S.; Faraonio, R.; Russo, G. Role of uL3 in Multidrug Resistance in p53-Mutated Lung Cancer Cells. Int. J. Mol. Sci. 2017, 18, 547. [Google Scholar] [CrossRef] [PubMed]
- Russo, A.; Russo, G. Ribosomal Proteins Control or Bypass p53 during Nucleolar Stress. Int. J. Mol. Sci. 2017, 18, 140. [Google Scholar] [CrossRef] [PubMed]
- Folkman, J.; Watson, K.; Ingber, D.; Hanahan, D. Induction of angiogenesis during the transition from hyperplasia to neoplasia. Nature 1989, 339, 58–61. [Google Scholar] [CrossRef] [PubMed]
- Carmeliet, P. Angiogenesis in life, disease and medicine. Nature 2005, 438, 932–936. [Google Scholar] [CrossRef] [PubMed]
- Vacca, A.; Ribatti, D. Angiogenesis and vasculogenesis in multiple myeloma: Role of inflammatory cells. Recent Results Cancer Res. 2011, 183, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Hose, D.; Moreaux, J.; Meissner, T.; Seckinger, A.; Goldschmidt, H.; Benner, A.; Mahtouk, K.; Hillengass, J.; Rème, T.; De Vos, J.; et al. Induction of angiogenesis by normal and malignant plasma cells. Blood 2009, 114, 128–143. [Google Scholar] [CrossRef] [PubMed]
- Vacca, A.; Ria, R.; Semeraro, F.; Merchionne, F.; Coluccia, M.; Boccarelli, A.; Scavelli, C.; Nico, B.; Gernone, A.; Battelli, F.; et al. Endothelial cells in the bone marrow of patients with multiple myeloma. Blood 2003, 102, 3340–3348. [Google Scholar] [CrossRef] [PubMed]
- Caligaris-Cappio, F.; Bergui, L.; Gregoretti, M.G.; Gaidano, G.; Gaboli, M.; Schena, M.; Zallone, A.Z.; Marchisio, P.C. Role of bone marrow stromal cells in the growth of human multiple myeloma. Blood 1991, 77, 2688–2693. [Google Scholar] [PubMed]
- Shafat, M.S.; Gnaneswaran, B.; Bowles, K.M.; Rushworth, S.A. The bone marrow microenvironment—Home of the leukemic blasts. Blood Rev. 2017, 31, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Lemaire, M.; Deleu, S.; De Bruyne, E.; Van Valckenborgh, E.; Menu, E.; Vanderkerken, K. The microenvironment and molecular biology of the multiple myeloma tumor. Adv. Cancer Res. 2011, 110, 19–42. [Google Scholar] [CrossRef] [PubMed]
- Hideshima, T.; Bergsagel, P.L.; Kuehl, W.M.; Anderson, K.C. Advances in biology of multiple myeloma: Clinical applications. Blood 2004, 104, 607–618. [Google Scholar] [CrossRef] [PubMed]
- Moyo, T.K.; Bouchnita, A.; Eymard, N.; Volpert, V.; Koury, M.J. Effects of bone marrow infiltration by multiple myeloma on erythropoiesis. Blood 2015, 126, 2143. [Google Scholar]
- Christoulas, D.; Terpos, E.; Dimopoulos, M.A. Pathogenesis and Management of Myeloma Bone Disease. Expert Rev. Hematol. 2009, 2, 385–398. [Google Scholar] [CrossRef] [PubMed]
- Silvestris, F.; Ciavarella, S.; De Matteo, M.; Tucci, M.; Dammacco, F. Bone-Resorbing Cells in Multiple Myeloma: Osteoclasts, Myeloma Cell Polykaryons, or Both? Oncologist 2009, 14, 264–275. [Google Scholar] [CrossRef] [PubMed]
- Merico, F.; Bergui, L.; Gregoretti, M.G.; Ghia, P.; Aimo, G.; Lindley, I.J.; Caligaris-Cappio, F. Cytokines involved in the progression of multiple myeloma. Clin. Exp. Immunol. 1993, 92, 27–31. [Google Scholar] [CrossRef] [PubMed]
- Pavlova, N.N.; Thompson, C.B. The emerging hallmarks of cancer metabolism. Cell Metab. 2016, 23, 27–47. [Google Scholar] [CrossRef] [PubMed]
- Brown, G.K. Glucose transporters: Structure, function and consequences of deficiency. J. Inherit. Metab. Dis. 2000, 23, 237–246. [Google Scholar] [CrossRef] [PubMed]
- Alfarouk, K.O.; Verduzco, D.; Rauch, C.; Muddathir, A.K.; Bashir, H.H.; Elhassan, G.O.; Ibrahim, M.E.; Orozco, J.D.P.; Cardone, R.A.; Reshkin, S.J. Glycolysis, tumor metabolism, cancer growth and dissemination. A new pH-based etiopathogenic perspective and therapeutic approach to an old cancer question. Oncoscience 2014, 1, 777–802. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J. Energy metabolism of cancer: Glycolysis versus oxidative phosphorylation. Oncol. Lett. 2012, 4, 1151–1157. [Google Scholar] [CrossRef] [PubMed]
- DeBerardinis, R.J.; Chandel, N.S. Fundamentals of cancer metabolism. Sci. Adv. 2016, 2, 2:1–2:18. [Google Scholar] [CrossRef] [PubMed]
- Pfeiffer, T.; Schuster, S.; Bonhoeffer, S. Cooperation and competition in the evolution of ATP-producing pathways. Science 2001, 292, 504–507. [Google Scholar] [CrossRef] [PubMed]
- Newsholme, P.; Procopio, J.; Lima, M.M.R.; Pithon-Curi, T.C.; Curi, R. Glutamine and glutamate—Their central role in cell metabolism and function. Cell Biochem. Funct. 2003, 21, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Pochini, L.; Scalise, M.; Galluccio, M.; Indiveri, C. Membrane transporters for the special amino acid glutamine: Structure/function relationships and relevance to human health. Front. Chem. 2014, 2, 61:1–61:23. [Google Scholar] [CrossRef] [PubMed]
- Altman, B.J.; Stine, Z.E.; Dang, C.V. From Krebs to clinic: Glutamine metabolism to cancer therapy. Nat. Rev. Cancer 2016, 16, 619–634. [Google Scholar] [CrossRef] [PubMed]
- Curthoys, N.P.; Watford, M. Regulation of glutaminase activity and glutamine metabolism. Annu. Rev. Nutr. 1995, 15, 133–159. [Google Scholar] [CrossRef] [PubMed]
- Moreadith, R.W.; Lehninger, A.L. The pathways of glutamate and glutamine oxidation by tumor cell mitochondria. Role of mitochondrial NAD(P)+-dependent malic enzyme. J. Biol. Chem. 1984, 259, 6215–6221. [Google Scholar] [PubMed]
- Hirschey, M.D.; DeBerardinis, R.J.; Diehl, A.M.; Drew, J.E.; Frezza, C.; Green, M.F.; Jones, L.W.; Ko, Y.H.; Le, A.; Lea, M.A.; et al. Dysregulated metabolism contributes to oncogenesis. Semin. Cancer Biol. 2015, 35, 129–150. [Google Scholar] [CrossRef] [PubMed]
- Lane, A.N.; Fan, T.W.M. Regulation of mammalian nucleotide metabolism and biosynthesis. Nucleic Acids Res. 2015, 43, 2466–2485. [Google Scholar] [CrossRef] [PubMed]
- Cory, J.G.; Cory, A.H. Critical roles of glutamine as nitrogen donors in purine and pyrimidine nucleotide synthesis: Asparaginase treatment in childhood acute lymphoblastic leukemia. In Vivo 2006, 20, 587–589. [Google Scholar] [PubMed]
- Alfarouk, K.O.; Shayoub, M.E.A.; Muddathir, A.K.; Elhassan, G.O.; Bashir, A.H.H. Evolution of Tumor Metabolism might Reflect Carcinogenesis as a Reverse Evolution process (Dismantling of Multicellularity). Cancers 2011, 3, 3002–3017. [Google Scholar] [CrossRef] [PubMed]
- Zu, X.L.; Guppy, M. Cancer metabolism: Facts, fantasy, and fiction. Biochem. Biophys. Res. Commun. 2004, 313, 459–465. [Google Scholar] [CrossRef] [PubMed]
- Gatenby, R.A.; Gillies, R.J. Why do cancers have high aerobic glycolysis? Nat. Rev. Cancer 2004, 4, 891–899. [Google Scholar] [CrossRef] [PubMed]
- Koppenol, W.H.; Bounds, P.L.; Dang, C.V. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat. Rev. Cancer 2011, 11, 325–337. [Google Scholar] [CrossRef] [PubMed]
- Fantin, V.R.; St-Pierre, J.; Leder, P. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell 2006, 9, 425–434. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Outschoorn, U.E.; Peiris-Pageès, M.; Pestell, R.G.; Sotgia, F.; Lisanti, M.P. Cancer metabolism: A therapeutic perspective. Nat. Rev. Clin. Oncol. 2017, 14, 11–31. [Google Scholar] [CrossRef] [PubMed]
- DeBerardinis, R.J.; Cheng, T. Q’s next: The diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene 2010, 29, 313–324. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Shi, X.; Zhang, L.; Yi, C.; Zhang, X.; Zhang, X. De novo glutamine synthesis: Importance for the proliferation of glioma cells and potentials for its detection with 13N-ammonia. Mol. Imaging 2016, 15, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Nicklin, P.; Bergman, P.; Zhang, B.; Triantafellow, E.; Wang, H.; Nyfeler, B.; Yang, H.; Hild, M.; Kung, C.; Wilson, C.; et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell 2009, 136, 521–534. [Google Scholar] [CrossRef] [PubMed]
- Thompson, C.B. Rethinking the regulation of cellular metabolism. Cold Spring Harb. Symp. Quant. Biol. 2011, 76, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Rathmell, J.C.; vander Heiden, M.G.; Harris, M.H.; Frauwirth, K.A.; Thompson, C.B. In the absence of extrinsic signals, nutrient utilization by lymphocytes is insufficient to maintain either cell size or viability. Mol. Cell 2000, 6, 683–692. [Google Scholar] [CrossRef]
- Glynn, S.A.; Albanes, D. Folate and cancer: A review of the literature. Nutr. Cancer 1994, 22, 101–119. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Phang, J.M. Oncogene and Cancer—From Bench to Clinic; InTech: London, UK, 2013; Volume 15, pp. 359–390. ISBN 978-953-51-0858-0. [Google Scholar]
- Nakano, A.; Miki, H.; Nakamura, S.; Harada, T.; Oda, A.; Amou, H.; Fujii, S.; Kagawa, K.; Takeuchi, K.; Ozaki, S.; et al. Up-regulation of hexokinaseII in myeloma cells: Targeting myeloma cells with 3-bromopyruvate. J. Bioenerg. Biomembr. 2012, 44, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Mathupala, S.P.; Ko, Y.H.; Pedersen, P.L. Hexokinase II: Cancer’s double-edged sword acting as both facilitator and gatekeeper of malignancy when bound to mitochondria. Oncogene 2006, 25, 4777–4786. [Google Scholar] [CrossRef] [PubMed]
- Robey, R.B.; Hay, N. Mitochondrial hexokinases, novel mediators of the antiapoptotic effects of growth factors and Akt. Oncogene 2006, 25, 4683–4696. [Google Scholar] [CrossRef] [PubMed]
- Lis, P.; Dyląg, M.; Niedźwiecka, K.; Ko, Y.H.; Pedersen, P.L.; Goffeau, A.; Ułaszewski, S. The HK2 dependent “Warburg Effect” and mitochondrial oxidative phosphorylation in cancer: Targets for effective therapy with 3-bromopyruvate. Molecules 2016, 21, 1730. [Google Scholar] [CrossRef] [PubMed]
- Azevedo-Silva, J.; Queirós, O.; Ribeiro, A.; Baltazar, F.; Young, K.H.; Pedersen, P.L.; Preto, A.; Casal, M. The cytotoxicity of 3-bromopyruvate in breast cancer cells depends on extracellular pH. Biochem. J. 2015, 467, 247–258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niedźwiecka, K.; Dyląg, M.; Augustyniak, D.; Majkowska-Skrobek, G.; Cal-Bąkowska, M.; Ko, Y.H.; Pedersen, P.L.; Goffeau, A.; Ułaszewski, S. Glutathione may have implications in the design of 3-bromopyruvate treatment protocols for both fungal and algal infections as well as multiple myeloma. Oncotarget 2016, 7, 65614–65626. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.D.; Deslandes, E.; Villedieu, M.; Poulain, L.; Duval, M.; Gauduchon, P.; Schwartz, L.; Icard, P. Effect of 2-deoxy-d-glucose on various malignant cell lines in vitro. Anticancer Res. 2006, 26, 3561–3566. [Google Scholar] [PubMed]
- Zhang, D.; Li, J.; Wang, F.; Hu, J.; Wang, S.; Sun, Y. 2-Deoxy-d-glucose targeting of glucose metabolism in cancer cells as a potential therapy. Cancer Lett. 2014, 355, 176–183. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Xia, J.; Xu, H.; Frech, I.; Tricot, G.; Zhan, F. NEK2 Promotes Aerobic Glycolysis in Multiple Myeloma Through Regulating Splicing of Pyruvate Kinase. J. Hematol. Oncol. 2017, 10, 17:1–17:11. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Wang, Y.; Liu, H.; Xu, X.; He, S.; Tang, J.; Huang, Y.; Miao, X.; Wu, Y.; Wang, Q. Pyruvate kinase isoform M2 (PKM2) participates in multiple myeloma cell proliferation, adhesion and chemoresistance. Leuk. Res. 2015, 39, 1428–1436. [Google Scholar] [CrossRef] [PubMed]
- Tamada, M.; Suematsu, M.; Saya, H. Pyruvate Kinase M2: Multiple faces for conferring benefits on cancer cells. Clin. Cancer Res. 2012, 18, 5554–5561. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, S.; Wada, N.; Kawano, Y.; Okuno, Y.; Kikukawa, Y.; Endo, S.; Nishimura, N.; Ueno, N.; Mitsuya, H.; Hata, H. Lactate, a putative survival factor for myeloma cells, is incorporated by myeloma cells through monocarboxylate transporters 1. Exp. Hematol. Oncol. 2015, 4, 12:1–12:8. [Google Scholar] [CrossRef] [PubMed]
- Sonveaux, P.; Végran, F.; Schroeder, T.; Wergin, M.C.; Verrax, J.; Rabbani, Z.N.; de Saedeleer, C.J.; Kennedy, K.M.; Diepart, C.; Jordan, B.F. Targeting lactate-fueled respiration selectively kills hypoxic tumor cells in mice. J. Clin. Investig. 2008, 118, 3930–3942. [Google Scholar] [CrossRef] [PubMed]
- Doherty, J.R.; Cleveland, J.L. Targeting lactate metabolism for cancer therapeutics. J. Clin. Investig. 2013, 123, 3685–3692. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, S.; Wada, N.; Kawano, Y.; Kikukawa, Y.; Mitsuya, H.; Hata, H. Lactate is a crucial energy source for multiple myeloma (MM) cells in bone marrow microenvironment. Blood 2013, 122, 3109. [Google Scholar]
- Zhang, S.; Hulver, M.W.; McMillan, R.P.; Cline, M.A.; Gilbert, E.R. The pivotal role of pyruvate dehydrogenase kinases in metabolic flexibility. Nutr. Metab. 2014, 11, 10:1–10:9. [Google Scholar] [CrossRef] [PubMed]
- Niewisch, M.R.; Kuçi, Z.; Wolburg, H.; Sautter, M.; Krampen, L.; Deubzer, B.; Handgretinger, R.; Bruchelt, G. Influence of dichloroacetate (DCA) on lactate production and oxygen consumption in neuroblastoma cells: Is DCA a suitable drug for neuroblastoma therapy? Cell. Physiol. Biochem. 2012, 29, 373–380. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, W.Y.; McGee, S.L.; Connor, T.; Mottram, B.; Wilkinson, A.; Whitehead, J.P.; Vuckovic, S.; Catley, L. Dichloroacetate inhibits aerobic glycolysis in multiple myeloma cells and increases sensitivity to bortezomib. Br. J. Cancer 2013, 108, 1624–1633. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, S.; Kawano, Y.; Yuki, H.; Okuno, Y.; Nosaka, K.; Mitsuya, H.; Hata, H. PDK1 inhibition is a novel therapeutic target in multiple myeloma. Br. J. Cancer 2013, 108, 170–178. [Google Scholar] [CrossRef] [PubMed]
- Romero-Garcia, S.; Moreno-Altamiranon, M.M.B.; Prado-Garcia, H.; Sánchez-García, F.J. Lactate contribution to the tumor microenvironment: Mechanisms, effects on immune cells and therapeutic relevance. Front. Immunol. 2016, 7, 52:1–52:11. [Google Scholar] [CrossRef] [PubMed]
- Rattigan, Y.I.; Patel, B.B.; Ackerstaff, E.; Sukenick, G.; Koutcher, J.A.; Glod, J.W.; Banerjee, D. Lactate is a mediator of metabolic cooperation between stromal carcinoma associated fibroblasts and glycolytic tumor cells in the tumor microenvironment. Exp. Cell Res. 2012, 318, 326–335. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, T.; Jimi, S.; Migita, K.; Takamatsu, Y.; Hara, S. Inhibition of glucose transporter 1 induces apoptosis and sensitizes multiple myeloma cells to conventional chemotherapeutic agents. Leuk. Res. 2016, 41, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Bolzoni, M.; Chiu, M.; Accardi, F.; Vescovini, R.; Airoldi, I.; Storti, P.; Todoerti, K.; Agnelli, L.; Missale, G.; Andreoli, R.; et al. Dependence on glutamine uptake and glutamine addiction characterize myeloma cells: A new attractive target. Blood 2016, 128, 667–679. [Google Scholar] [CrossRef] [PubMed]
- Accardi, F.; Chiu, M.; Bolzoni, M.; Storti, P.; Todoerti, K.; Agnelli, L.; Ferrari, M.; Missale, G.; Aversa, F.; Bussolati, O.; et al. Ammonium Production and Glutamine-Addiction of Myeloma Cells: New Attractive Targets in Multiple Myeloma. Blood 2014, 124, 2067. [Google Scholar]
- Minetto, P.; Soncini, D.; Cagnetta, A.; Guolo, F.; Retali, V.; Rivoli, G.; Bisso, N.; Di Felice, N.; Miglino, M.; Canepa, L.; et al. Glutamine-Dependence Targeting By Asparaginase Significantly Increases Anti-Myeloma Activity of Proteasome Inhibitors. Blood 2017, 130, 1796. [Google Scholar]
- Giuliani, N.; Chiu, M.; Bolzoni, M.; Accardi, F.; Bianchi, M.G.; Toscani, D.; Aversa, F.; Bussolati, O. The potential of inhibiting glutamine uptake as a therapeutic target for multiple myeloma. Expert Opin. Ther. Targets 2017, 21, 231–234. [Google Scholar] [CrossRef] [PubMed]
- Corbet, C.; Feron, O. Metabolic and mind shifts: From glucose to glutamine and acetate addictions in cancer. Curr. Opin. Clin. Nutr. Metab. Care 2015, 18, 346–353. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Cui, H. Targeting Glutamine Induces Apoptosis: A Cancer Therapy Approach. Int. J. Mol. Sci. 2015, 16, 22830–22855. [Google Scholar] [CrossRef] [PubMed]
- Jeon, Y.J.; Khelifa, S.; Ratnikov, B.; Scott, D.A.; Feng, Y.; Parisi, F.; Ruller, C.; Lau, E.; Kim, H.; Brill, L.M.; et al. Regulation of glutamine carrier proteins by RNF5 determines breast cancer response to ER stressinducing chemotherapies. Cancer Cell 2015, 27, 354–369. [Google Scholar] [CrossRef] [PubMed]
- Gabay, M.; Li, Y.; Felsher, D.W. MYC activation is a hallmark of cancer initiation and maintenance. Cold Spring Harb. Perspect. Med. 2014, 4, 6:1–6:13. [Google Scholar] [CrossRef] [PubMed]
- Effenberger, M.; Bommert, K.S.; Kunz, V.; Kruk, J.; Leich, E.; Rudelius, M.; Bargou, R.; Bommert, K. Glutaminase inhibition in multiple myeloma induces apoptosis via MYC degradation. Oncotarget 2017, 8, 85858–85867. [Google Scholar] [CrossRef] [PubMed]
- Wise, D.R.; DeBerardinis, R.J.; Mancuso, A.; Sayed, N.; Zhang, X.Y.; Pfeiffer, H.K.; Nissim, I.; Daikhin, E.; Yudkoff, M.; McMahon, S.B.; et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc. Natl. Acad. Sci. USA 2008, 105, 18782–18787. [Google Scholar] [CrossRef] [PubMed]
- Gonsalves, W.I.; Ramakrishnan, V.; Hitosugi, T.; Ghosh, T.; Jevremovic, D.; Dutta, T.; Sakrikar, D.; Petterson, X.M.; Wellik, L.; Kumar, S.K.; et al. Glutamine-derived 2-hydroxyglutarate is associated with disease progression in plasma cell malignancies. JCI Insight 2018, 3, e94543. [Google Scholar] [CrossRef] [PubMed]
- Casey, S.C.; Tong, L.; Li, Y.; Do, R.; Walz, S.; Fitzgerald, K.N.; Gouw, A.M.; Baylot, V.; Gütgemann, I.; Eilers, M.; et al. MYC regulates the antitumor immune response through CD47 and PD-L1. Science 2016, 352, 227–231. [Google Scholar] [CrossRef] [PubMed]
- Cacace, A.; Sboarina, M.; Vazeille, T.; Sonveaux, P. Glutamine activates STAT3 to control cancer cell proliferation independently of glutamine metabolism. Oncogene 2017, 36, 2074–2084. [Google Scholar] [CrossRef] [PubMed]
- Roland, C.L.; Arumugam, T.; Deng, D.; Liu, S.H.; Philip, B.; Gomez, S.; Burns, W.R.; Ramachandran, V.; Wang, H.; Cruz-Monserrate, Z.; et al. Cell surface lactate receptor GPR81 is crucial for cancer cell survival. Cancer Res. 2014, 74, 5301–5310. [Google Scholar] [CrossRef] [PubMed]
- Yao, C.; Li, Y.Y.; Li, J.; Zhang, H.Y.; Wang, F.; Bai, X.; Li, S.S. STAT3 regulates hypoxia-induced epithelial mesenchymal transition in oesophageal squamous cell cancer. Oncol. Rep. 2016, 36, 108–116. [Google Scholar] [CrossRef]
- Denko, N.C. Hypoxia, HIF1 and glucose metabolism in the solid tumour. Nat. Rev. Cancer 2008, 8, 705–713. [Google Scholar] [CrossRef] [PubMed]
- Maiso, P.; Huynh, D.; Moschetta, M.; Sacco, A.; Aljawai, Y.; Mishima, Y.; Asara, J.M.; Roccaro, A.M.; Kimmelman, A.C.; Ghobrial, I.M. Metabolic signature identifies novel targets for drug resistance in Multiple Myeloma. Cancer Res. 2015, 75, 2071–2082. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, C.F.; Anthony, K.; Krupa, S.; Buchoff, J.; Day, M.; Hannay, T.; Buetow, K.H. PID: The Pathway Interaction Database. Nucleic Acids Res. 2009, 37, D674–679. [Google Scholar] [CrossRef] [PubMed]
- Mulligan, G.; Mitsiades, C.; Bryant, B.; Zhan, F.; Chng, W.J.; Roels, S.; Koenig, E.; Fergus, A.; Huang, Y.; Richardson, P.; et al. Gene expression profiling and correlation with outcome in clinical trials of the proteasome inhibitor bortezomib. Blood 2007, 109, 3177–3188. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Jia, X.; Duan, Y.; Xiao, H.; Sundqvist, K.G.; Permert, J.; Wang, F. Excess glucose induces hypoxia-inducible factor-1α in pancreatic cancer cells and stimulates glucose metabolism and cell migration. Cancer Biol. Ther. 2013, 14, 428–435. [Google Scholar] [CrossRef] [PubMed]
- Zub, K.A.; de Sousa, M.M.L.; Sarno, A.; Sharma, A.; Demirovic, A.; Rao, S.; Young, C.; Aas, P.A.; Ericsson, I.; Sundan, A.; et al. Modulation of Cell Metabolic Pathways and Oxidative Stress Signaling Contribute to Acquired Melphalan Resistance in Multiple Myeloma Cells. PLoS ONE 2015, 10, 3:1–3:20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, X.; Fang, L.; Gibbs, S.; Huang, Y.; Dai, Z.; Wen, P.; Zheng, X.; Sadee, W.; Sun, D. Glucose uptake inhibitor sensitizes cancer cells to daunorubicin and overcomes drug resistance in hypoxia. Cancer Chemother. Pharmacol. 2007, 59, 495–505. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Bajpai, R.; Sharma, H.; Heitmeier, M.; Jain, A.D.; Matulis, S.M.; Nooka, A.K.; Mishra, R.K.; Hruz, P.W.; Schiltz, G.E.; et al. Development of GLUT4-selective antagonists for multiple myeloma therapy. Eur. J. Med. Chem. 2017, 139, 573–586. [Google Scholar] [CrossRef] [PubMed]
- Bajpai, R.; Matulis, S.M.; Wei, C.; Nooka, A.K.; Von Hollen, H.E.; Lonial, S.; Boise, L.H.; Shanmugam, M. Targeting glutamine metabolism in multiple myeloma enhances BIM binding to BCL-2 eliciting synthetic lethality to venetoclax. Oncogene 2016, 35, 3955–3964. [Google Scholar] [CrossRef] [PubMed]
- Dalva-Aydemir, S.; Bajpai, R.; Martinez, M.; Adekola, K.U.; Kandela, I.; Wei, C.; Singhal, S.; Koblinski, J.E.; Raje, N.S.; Rosen, S.T.; et al. Targeting the Metabolic Plasticity of Multiple Myeloma with FDA-Approved Ritonavir and Metformin. Clin. Cancer Res. 2015, 21, 1161–1171. [Google Scholar] [CrossRef] [PubMed]
- Kurtoglu, M.; Gao, N.; Shang, J.; Maher, J.C.; Lehrman, M.A.; Wangpaichitr, M.; Savaraj, N.; Lane, A.N.; Lampidis, T.J. Under normoxia, 2-deoxy-D-glucose elicits cell death in select tumor types not by inhibition of glycolysis but by interfering with N-linked glycosylation. Mol. Cancer Ther. 2007, 6, 3049–3058. [Google Scholar] [CrossRef] [PubMed]
- Maher, J.C.; Krishan, A.; Lampidis, T.J. Greater cell cycle inhibition and cytotoxicity induced by 2-deoxy-D-glucose in tumor cells treated under hypoxic vs aerobic conditions. Cancer Chemother. Pharmacol. 2004, 53, 116–122. [Google Scholar] [CrossRef] [PubMed]
- Miao, P.; Sun, X.; Liu, J.; Huang, G. Lactate dehydrogenase a in cancer: A promising target for diagnosis and therapy. IUBMB Life 2013, 65, 904–910. [Google Scholar] [CrossRef] [PubMed]
- Mattaini, K.R.; Sullivan, M.R.; Vander Heiden, M.G. The importance of serine metabolism in cancer. J. Cell Biol. 2016, 214, 249–257. [Google Scholar] [CrossRef] [PubMed]
- Locasale, J.W. Serine, glycine and one-carbon units: Cancer metabolism in full circle. Nat. Rev. Cancer 2013, 13, 572–583. [Google Scholar] [CrossRef] [PubMed]
- Zaal, E.A.; Wu, W.; Jansen, G.; Zweegman, S.; Cloos, J.; Berkers, C.R. Bortezomib resistance in multiple myeloma is associated with increased serine synthesis. Cancer Metab. 2017, 5, 7:1–7:12. [Google Scholar] [CrossRef] [PubMed]
- Maddocks, O.D.K.; Athineos, D.; Cheung, E.C.; Lee, P.; Zhang, T.; van den Broek, N.J.F.; Mackay, G.M.; Labuschagne, C.F.; Gay, D.; Kruiswijk, F.; et al. Modulating the therapeutic response of tumours to dietary serine and glycine starvation. Nature 2017, 544, 372–376. [Google Scholar] [CrossRef] [PubMed]
- Patra, K.C.; Hay, N. The pentose phosphate pathway and cancer. Trends Biochem. Sci. 2014, 39, 347–354. [Google Scholar] [CrossRef]
- Horecker, B.L. The pentose phosphate pathway. J. Biol. Chem. 2002, 277, 47965–47971. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Huang, R.; Ding, J.; Ji, D.; Song, B.; Yuan, L.; Chang, H.; Chen, G. Multiple myeloma acquires resistance to EGFR inhibitor via induction of pentose phosphate pathway. Sci. Rep. 2015, 5, 9925:1–9925:8. [Google Scholar] [CrossRef] [PubMed]
- Normanno, N.; De Luca, A.; Bianco, C.; Strizzi, L.; Mancino, M.; Maiello, M.R.; Carotenuto, A.; de Feo, G.; Caponigro, F.; Salomon, D.S. Epidermal growth factor receptor (EGFR) signaling in cancer. Gene 2006, 366, 2–16. [Google Scholar] [CrossRef] [PubMed]
- Hothersall, J.S.; Gordge, M.; Noronha-Dutra, A.A. Inhibition of NADPH supply by 6-aminonicotinamide: Effect on glutathione, nitric oxide and superoxide in J774 cells. FEBS Lett. 1998, 434, 97–100. [Google Scholar] [CrossRef]
- Thompson, R.M.; Dytfeld, D.; Reyes, L.; Robinson, R.M.; Smith, B.; Manevich, Y.; Jakubowiak, A.; Komarnicki, M.; Przybylowicz-Chalecka, A.; Szczepaniak, T.; et al. Glutaminase inhibitor CB-839 synergizes with carfilzomib in resistant multiple myeloma cells. Oncotarget 2017, 8, 35863–35876. [Google Scholar] [CrossRef] [PubMed]
- Katz, B.Z. Adhesion molecules—The lifelines of multiple myeloma cells. Semin. Cancer Biol. 2010, 20, 186–195. [Google Scholar] [CrossRef] [PubMed]
- Burger, J.A.; Ghia, P.; Rosenwald, A.; Caligaris-Cappio, F. The microenvironment in mature B-cell malignancies: A target for new treatment strategies. Blood 2009, 114, 3367–3375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Damiano, J.S.; Cress, A.E.; Hazlehurst, L.A.; Shtil, A.A.; Dalton, W.S. Cell adhesion mediated drug resistance (CAM-DR): Role of integrins and resistance to apoptosis in human myeloma cell lines. Blood 1999, 93, 1658–1667. [Google Scholar] [PubMed]
- Zhu, J.; Wang, M.; Cao, B.; Hou, T.; Mao, X. Targeting the phosphatidylinositol 3-kinase/AKT pathway for the treatment of multiple myeloma. Curr. Med. Chem. 2014, 21, 3173–3187. [Google Scholar] [CrossRef] [PubMed]
- Steinbrunn, T.; Stühmer, T.; Sayehli, C.; Chatterjee, M.; Einsele, H.; Bargou, R.C. Combined targeting of MEK/MAPK and PI3K/Akt signalling in multiple myeloma. Br. J. Haematol. 2012, 159, 430–440. [Google Scholar] [CrossRef] [PubMed]
- Medina, E.A.; Oberheu, K.; Polusani, S.R.; Ortega, V.; Velagaleti, G.V.; Oyajobi, B.O. PKA/AMPK signaling in relation to adiponectin’s antiproliferative effect on multiple myeloma cells. Leukemia 2014, 28, 2080–2089. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Lin, L.; Cao, L.; Zhang, X.; Song, X.; Hao, J.; Zhang, Y.; Wei, R.; Huang, X.; Lu, J.; et al. Extracellular matrix protein Reelin promotes myeloma progression by facilitating tumor cell proliferation and glycolysis. Sci. Rep. 2017, 7, 45305:1–45305:12. [Google Scholar] [CrossRef] [PubMed]
- Tamura, H. Immunopathogenesis and immunotherapy of multiple myeloma. Int. J. Hematol. 2018, 107, 278–285. [Google Scholar] [CrossRef] [PubMed]
- Krejcik, J.; Frerichs, K.A.; Nijhof, I.S.; van Kessel, B.; van Velzen, J.F.; Bloem, A.C.; Broekmans, M.E.C.; Zweegman, S.; van Meerloo, J.; Musters, R.J.P. Monocytes and Granulocytes Reduce CD38 Expression Levels on Myeloma Cells in Patients Treated with Daratumumab. Clin. Cancer Res. 2017, 23, 7498–7511. [Google Scholar] [CrossRef] [PubMed]
- Hosen, N.; Matsunaga, Y.; Hasegawa, K.; Matsuno, H.; Nakamura, Y.; Makita, M.; Watanabe, K.; Yoshida, M.; Satoh, K.; Morimoto, S. The activated conformation of integrin β7 is a novel multiple myeloma-specific target for CAR T cell therapy. Nat. Med. 2017, 23, 1436–1443. [Google Scholar] [CrossRef] [PubMed]
- Kouidhi, S.; Ben Ayed, F.; Benammar Elgaaied, A. Targeting Tumor Metabolism: A New Challenge to Improve Immunotherapy. Front. Immunol. 2018, 9, 353. [Google Scholar] [CrossRef] [PubMed]
- Beckermann, K.E.; Dudzinski, S.O.; Rathmell, J.C. Dysfunctional T cell metabolism in the tumor microenvironment. Cytokine Growth Factor Rev. 2017, 35, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Noël, G.; Fontsa, M.L.; Willard-Gallo, K. The impact of tumor cell metabolism on T cell-mediated immune responses and immuno-metabolic biomarkers in cancer. Semin. Cancer Biol. 2018, 1–28. [Google Scholar] [CrossRef] [PubMed]
- Housman, G.; Byler, S.; Heerboth, S.; Lapinska, K.; Longacre, M.; Snyder, N.; Sarkar, S. Drug resistance in cancer: An overview. Cancers 2014, 6, 1769–1792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parkin, B.; Ouillette, P.; Li, Y.; Keller, J.; Lam, C.; Roulston, D.; Li, C.; Shedden, K.; Malek, S.N. Clonal evolution and devolution after chemotherapy in adult acute myelogenous leukemia. Blood 2013, 121, 369–377. [Google Scholar] [CrossRef] [PubMed]
- Navin, N.; Krasnitz, A.; Rodgers, L.; Cook, K.; Meth, J.; Kendall, J.; Riggs, M.; Eberling, Y.; Troge, J.; Grubor, V.; et al. Inferring tumor progression from genomic heterogeneity. Genome Res. 2010, 20, 68–80. [Google Scholar] [CrossRef] [PubMed]
 , reduction;
, reduction;  , strong reduction;
, strong reduction;  , increase;
, increase;  , strong increase.
, strong increase.
 , reduction;
, reduction;  , strong reduction;
, strong reduction;  , increase;
, increase;  , strong increase.
, strong increase.
| Drug | Target | Resistance Effect in MM | Combination Treatment to Lower Resistance |
|---|---|---|---|
| Bortezomib | Proteasome inhibitor | HKII  | LDHA knockdown |
| Daunorubicin | DNA-RNA synthesis inhibitor | Glucose metabolism  | Phloretin |
| Gefitinib | EGFR inhibitor | PPP  | 6-AN |
| Afatinib | EGFR inhibitor | PPP  | Unknown |
| CB-839 | GLS inhibitor | Glutaminolysis  | Proteasome inhibitors (bortezomib, carfilzomib, ixazomib, oprozomib) |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
El Arfani, C.; De Veirman, K.; Maes, K.; De Bruyne, E.; Menu, E. Metabolic Features of Multiple Myeloma. Int. J. Mol. Sci. 2018, 19, 1200. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19041200
El Arfani C, De Veirman K, Maes K, De Bruyne E, Menu E. Metabolic Features of Multiple Myeloma. International Journal of Molecular Sciences. 2018; 19(4):1200. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19041200
Chicago/Turabian StyleEl Arfani, Chaima, Kim De Veirman, Ken Maes, Elke De Bruyne, and Eline Menu. 2018. "Metabolic Features of Multiple Myeloma" International Journal of Molecular Sciences 19, no. 4: 1200. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19041200