1. Introduction
Surface meshes are typically used in modeling, animation, numerical simulation, and many other applications. Molecular surface meshes play a vital role in the study of evolution and interaction of molecules and in measuring their areas and volumes [
1,
2]. These meshes are used in various fields of computational biology, such as protein folding, structure prediction, docking and implicit-solvent modeling. However, these meshes are generated in raw form, thereby containing low-quality elements. Such raw meshes with low-quality elements are difficult to be directly used in downstream applications. Recent development in mathematical modeling and simulation of biomolecules, especially in implicit solvent modeling, demands a proper refinement of these molecular meshes to remove low-quality elements and improve meshing quality [
3].
The molecular Gaussian surface is represented by the blending of a set of Gaussian functions. Various algorithms have been developed to triangulate and render molecular surfaces. TMSmesh [
4,
5] is a manifold triangular meshing framework used for meshing large Gaussian molecular surfaces. TMSmesh can handle a number of tasks, such as overlapping, gap filling, and seed selection, that need to be considered in traditional continuation methods. It succeeds in surface mesh generation for biomolecules containing more than one million atoms. TMSmesh generates meshes with satisfactory quality in terms of uniformity and manifoldness.The output mesh preserves the geometry and features (topology, area and volume, and local curvature) of the input mesh. However, the computational efficiency still needs to be improved. In this regard, Liu et al. [
6] proposed an improved version i.e., TMSmesh 2.0. Their results show that TMSmesh 2.0 is robust, efficient, and more than thirty times faster as compared to the previous version [
4].
Quality requirements in mesh processing which defines the class of acceptable and supported models varies from application to application [
7]. Molecular remeshing has its own challenges and issues, such as raw input meshes, very small angles, and complex geometry. Different methods have been proposed to handle various issues in molecular surface remeshing. ISO2mesh [
8] is a mesh processing toolbox used for mesh smoothing and tetrahedral mesh generation. It can be used for molecular mesh smoothing, but defects such as self-intersecting triangles and small angle triangles remain in its results. Recently, Liu et al. [
9] proposed a method called SMOPT to improve molecular remeshing with the removal of redundant vertices and self-intersecting triangles. However, there is no considerable improvement in minimal and maximal angles in SMOPT results. The literature study shows that the existing methods for molecular refreshing somehow fail to handle maximal and minimal angles, self-intersecting triangles, redundant vertices, and other defects. Therefore, efficient methods for quality improvement of molecular meshes are demanded.
In this paper, we proposed a simple method to improve molecular surface meshes. The proposed method starts with real-time adaptive remeshing (RAR) [
10] initialization followed by aspect ratio improvement. Furthermore, a cut-and-fill method is applied to remove invalid regions and refill them. The newly filled regions are further improved via edge split, edge collapse, and vertex translation with the condition of minimal angle improvement. We compared our results with two recent methods including SMOPT and ISO2mesh and found a significant improvement in the meshing quality. The results reveal that our method preserves the volume and area of the input mesh, and has a solvation energy similar to SMOPT, removes redundant vertices, and eliminates small angles (i.e., <30
). The main contributions of this study are as follows:
We propose a mesh refinement method for smoothing molecular surface meshes and improve the minimal and maximal angles to an angle bound [30, 120].
A cut-and-fill strategy is used to carefully remove the invalid regions with redundant vertices and/or small angles.
A global smoothing method is used to improve aspect ratio, and local operators are used to refine the newly filled holes and improve the minimal angle.
2. Related Work
Molecular surfaces have various definitions based on their molecular structure. In a recent study [
11], Chen and Lu summarized molecular surfaces, including van der Waals (VDW) surfaces, solvent accessible surfaces (SASs), solvent excluded surfaces (SESs), molecular skin surfaces, minimal molecular surfaces, and Gaussian surfaces. In this section, we briefly review the existing methods in molecular surface meshing. We recommend books [
12] and review articles [
11,
13,
14] for detailed studies. Alliez et al. [
13] reviewed surface remeshing techniques generally used in computer graphics and geometry processing applications. Chen and Lu [
11] conducted a review specific in molecular surface remeshing. Similarly, Bade et al. [
14] compared state-of-the-art methods of medical mesh smoothing.
In computer graphics, numerous surface remeshing methods have been proposed. These methods can be classified as mesh-simplification-based methods [
15,
16], Delaunay insertion methods [
17], advancing-front-based methods [
18], field-based approaches [
19,
20], and local-operator-based mesh optimization [
10,
21]. In addition, global optimization methods which include parametrization-based methods [
22,
23], discrete clustering [
24], and direct 3D optimization [
25,
26,
27,
28] are also available. Furthermore, segmentation-based meshing are also used, where input meshes are segmented prior to remeshing, which helps to preserve sharp features [
29,
30]. In terms of implicit feature preservation, several approaches exhibit efficient feature functions [
24,
31]. Laplacian smoothing [
32] is the simplest method, which involves moving each vertex to the central position of its neighbor. Equation (
1) computes the new position
for a free vertex
as the median of the positions of the
n vertices
in its one-ring neighborhood.
Taubin [
33] proposed a LowPass filter, which combines two Laplace like filters, one with positive weight and one with negative weight. The Taubin method [
33] computes new position
from old position
using Equation (
2):
Here the weighting factor
is commonly used as
.
is the weighting factor, which is replaced by another weighting factor
with a small value
.
is used to set the value of
a bit smaller than
. These two weighting factors including
and
are alternatively applied for backward translation [
33].
Recent methods show a significant improvement in minimal and maximal angles and meshing quality for graphical models. For example, centroidal Voronoi tessellation (CVT) [
34,
35] smooths meshes by translating vertices to new positions which optimize an energy function. Real-time adaptive remeshing (RAR) [
10] is a high-quality adaptive remeshing approach that is suitable for real-time applications. Mansouri and Ebrahimnezhad proposed a curvature-adapted subdivision method [
36], which minimizes the distortion error and improves the aspect ratio (AR). Yan and Wonka [
37] proposed a non-obtuse remeshing method using additional operators with CVT to avoid short Voronoi edges. In this manner, they remove small angles (<30
) and obtuse angles. However, for noisy meshes, their method fails to achieve the desired angle bound (i.e., [30
, 90
]). Recently, Hu et al. [
31] proposed a remeshing method with common local edge-based operators (edge split, edge collapse, and edge flip and edge split) and vertex smoothing. They generated results with a minimal angle higher than 35
with feature preservation. These approaches can efficiently handle common graphical models, such as CAD models and man-made objects. However, for molecular remesing, these methods are not directly applicable. Tiny triangles (with zero or near-zero degree angles), redundant vertices, feature preservation, and complex geometry are the specific challenges with molecular surface meshes where the existing remeshing methods often fail.
The removal of defects from a raw mesh is also an interesting topic in mesh processing. Ju presented PolyMender [
38], a simple yet robust method for repairing polygonal models. The algorithm generates closed surface meshes, repairing all existing defects in the input model with features preservation. MeshFix [
39] is another tool used to convert raw digitized polygons to clean mesh avoiding holes, non-manifold elements, and degenerate or intersecting elements. Experiments show that MeshFix provides results that have a higher visual quality, are more accurate, and add fewer new triangles compared to their previous methods. Attene et al. [
7] summarized typical defects that make a 3D model unsuitable for different applications and reviewed the existing refreshing techniques.
Meshlab [
40] is another tool used for surface repair, reconstruction, and smoothing purposes. It enables the implementation of several state-of-the-art methods such as mesh smoothing methods [
33,
41,
42,
43] and several modules for mesh cleaning and repairing. Similarly, Graphite [
44] is also used for surface smoothing, remeshing, 3D modeling, and surface repairing purposes. It is used to visualize holes and non-manifold configurations, to fill holes, and to remove self-intersections.
In the molecular modeling community, Decherchi and Rocchia [
45] proposed a ray-casting method for the triangulation of complex manifold surfaces in the nano-bioscience field. They summarized various applications of molecular surfaces in implicit solvent modeling and simulations using the boundary element method (BEM) and the finite element method (FEM). TMSmesh [
4,
5] generates molecular surface meshes for molecules. TMSmesh 2.0 can efficiently generate manifold surface meshes for biomolecules with more than one million atoms with shape and feature preservations [
46]. The Molecular Finite Element Solver (mFES) [
47] is a tool that uses tetrahedral finite elements to calculate electrostatic potentials of large molecular systems.
ISO2mesh [
8] is a free Matlab/octave-based toolbox used for mesh generation and processing. It is used to create tetrahedral meshes from surface meshes and 3D binary and gray-scale volumetric images such as segmented MRI/CT scans. ISO2mesh is also used for molecular mesh smoothing, but it fails to handle self-intersecting triangle pairs and small angle triangles. Liu et al. [
9] proposed an algorithm called SMOPT for molecular surface remeshing. They used local modifications on the mesh to improve the mesh quality, eliminate redundant vertices, avoid non-manifold errors, and remove intersecting triangles. For mesh smoothing, SMOPT has improved Laplacian smoothing, which is given in Equation (
3).
where
is the parameter to control the rate of smoothing,
represents the number of vertices in the one ring, and
represent the
jth adjacent vertex in the one ring of the
ith vertex. SMOPT results show a significant improvement in the mesh quality. Still, there are very small angles which destroy the quality of the triangles.
4. The Proposed Molecular Remeshing Method
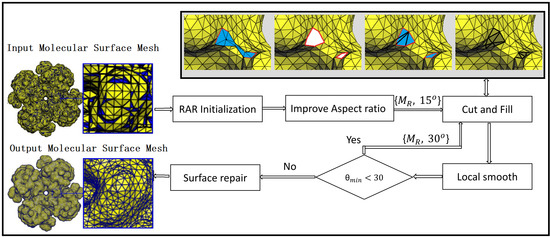
Our method starts with RAR remeshing followed by aspect ratio improvement. After this, local operators cut, fill, and smooth are repeated until a high-quality mesh is generated. Algorithm 1 represents the main modules of our method, which is described below. Here represents input mesh, represents the intermediate mesh, and is the output mesh.
| Algorithm 1 Molecular Remeshing () |
- 1:
- 2:
// Algorithm 2 - 3:
- 4:
- 5:
// Algorithm 3 - 6:
- 7:
- 8:
// Algorithm 3 - 9:
- 10:
- 11:
|
Figure 2 shows the pipeline of the proposed method. In the first step, the input model is remeshed with RAR. In the second step, the aspect ratios of the mesh are improved using Algorithm 2. In the third step, the local regions around the minimal angle with threshold
= 15
are removed. In the next step, the holes created are refilled. In the fifth step, the newly filled regions are smoothed locally (Algorithm 3). Similarly, these last three steps (i.e., cut, fill, and smooth) are again repeated with threshold
= 30
. Finally, the mesh is passed through a surface repair module to avoid any intersecting faces or non-manifoldness (if any). The algorithm is further described in the following subsections.
4.1. Initialization with RAR Method
The input mesh has many zero degree angles and redundant vertices. Initially, we apply the RAR method to improve the input mesh. We select RAR [
10] for remeshing because this method is comparatively easy to control, simple to implement, and computationally efficient. RAR uses Equation (
8) as an adaptive sizing function to compute the edge length
for an edge
with vertices
and
at its two ends.
where the sizing field
is calculated for vertex
using Equation (
9):
where
is error tolerance, and
is the maximum absolute curvature of a vertex. The maximum absolute curvature
for a vertex
is calculated from the mean curvature
and Gaussian curvature
using Equations (
10)–(
12) [
52]:
Here, represents the adjacent triangle angles around a vertex , and represents the corresponding Voronoi area.
An edge with shorter actual length than collapses and splits if its actual length is longer than . The RAR method improves the quality for most of the triangles, but it fails to remove all zero degree angles from a raw input molecular mesh and we need further improvements.
4.2. Aspect Ratio Improvement
After pre-processing with RAR we use a global smoothing method to improve the aspect ratios of the triangles. Algorithm 2 improves the aspect ratios and is described below.
| Algorithm 2 Improve Aspect Ratio () |
- 1:
for each vertex do - 2:
- 3:
if then - 4:
// Translate the vertex - 5:
end if - 6:
end for - 7:
- 8:
|
For each vertex, we calculate the new position as the center of mass of the corresponding Voronoi cell in the same manner as CVT [
53] and then as the Laplacian center (Equation (
1)) of the one-ring neighbourhood. The Laplacian method [
32] is used for computing the new position
as the median position of its one-ring neighbors using Equation (
1).
Equation (
13) computes aspect ratio for vertex
v as the average of the aspect ratios of the adjacent triangles. Equation (
14) computes aspect ratio for a single triangle
t. The aspect ratio is computed for vertex new position as well as its original position using Equation (
13). The vertex is translated to the new position if it improves the aspect ratio, otherwise translation is skipped. This process of aspect ratio improvement is repeated on all vertices of the mesh.
where
T represents the triangles set in the one-ring neighborhood of vertex
v, and
N represents the number of triangles in the one-ring neighborhood of vertex
v, whereas
a,
b, and
c are the lengths of the triangle’s edges and
.
4.3. The Cut-and-Fill Strategy
The input mesh has a number of redundant vertices and very tiny angle triangles. The mesh smoothing methods and the edge based operations (edge splitting, edge collapsing, and edge flipping) fail to handle these tiny angles. Therefore, we use a cut & fill strategy to handle this issue.
Figure 3 shows the invalid regions in a mesh to be cut and refilled.
Each triangle with an angle
is labeled as a small triangle. The one-ring neighborhood around each vertex of a small triangle is included in the local regions for the cut-and-fill operations. Each small triangle with its local region around is removed and the holes are refilled (see
Figure 4 and
Figure 5). Small holes are filled by connecting the boundary edges of the hole, whereas for filling large holes new vertices are also inserted inside the hole. The boundary vertices of each hole are stored in a queue. For the four adjacent vertices
,
,
, and
, we select from center vertex
. The side vertices
and
are connected with each other if the minimal angle for the new triangle is less than 15
; otherwise, the connection is skipped. The same step is repeated for connecting
and
. If the connection is not possible, the vertex is changed, and the process is repeated again. In other words, each
ith vertex of the boundary vertices is connected with the
th vertex if it does not create angle smaller than 15
. If an edge is larger than
, where
represents the average edge length calculated from the triangles adjacent to the hole. Edge splitting in the later local smoothing also creates new vertices inside the holes. Small tiny regions are created during the mesh processing which are removed with the cut-and-fill method.
Figure 5 shows a narrow region of tiny triangles connected at two sides of the surface mesh (blue color). The narrow region is removed making two holes on both sides. The holes are filled and smoothed independently.
4.4. Local Smooth
The cut-and-fill strategy eliminates tiny triangles and redundant vertices. However, the newly filled regions still need smoothing for further improvements of the minimal angle. Unlike RAR, our method has a local smoothing module to improve the minimal angles locally. Our method can eliminate all small angles, as shown in
Section 5, whereas the RAR method does not eliminate all angles smaller than 30
. Algorithm 3 smooths the newly filled regions locally. In the following, we describe the local smoothing method of the algorithm.
| Algorithm 3 Smooth New Filled Regions () |
- 1:
Collapse() - 2:
Split() - 3:
for each vertex do - 4:
- 5:
for do - 6:
- 7:
if (Vertex translation improves minimal angle) then - 8:
// Translate the vertex - 9:
end if - 10:
end for - 11:
end for - 12:
- 13:
|
In the first step, we collapse the short edges (an edge with opposite angle <30) to remove small angles. In the second step, we split the long edges (an edge with opposite angle >90). The remaining steps are applied to each vertex in the newly filled regions. In the fourth step, the Laplacian center of the one ring around the vertex is calculated. Each smoothing iteration calculates the new position near the Laplacian center . Here, represents step size, which is a small distance (to be added with ), whereas k is the direction of the vertex translation. The direction k is randomly selected to calculate the new position at a distance from the vertex’s current position, which could be toward the left, right, up, or down side with respect to the current position. In each iteration, the vertex is translated to the new position , if it improves the minimal angle.
4.5. Surface Repair
We have used constraints for applying meshing operators, such as minimal angle improvements and aspect ratio improvements, which prohibit meshes from creating new defects. However, due to complex structures of molecular meshes, some defects may still exist, causing failure in volume mesh generation by TetGen. Here, we used the “surface repair” module from Graphite [
44] to avoid self-intersection and other possible defects.