Berberine Protects against NEFA-Induced Impairment of Mitochondrial Respiratory Chain Function and Insulin Signaling in Bovine Hepatocytes
Abstract
:1. Introduction
2. Results
2.1. Blood Biochemical Index and Liver Steatosis in Dairy Cows
2.2. Dairy Cows with Fatty Liver Displayed Impaired Hepatic Insulin Signaling and Mitochondrial Respiratory Chain Function
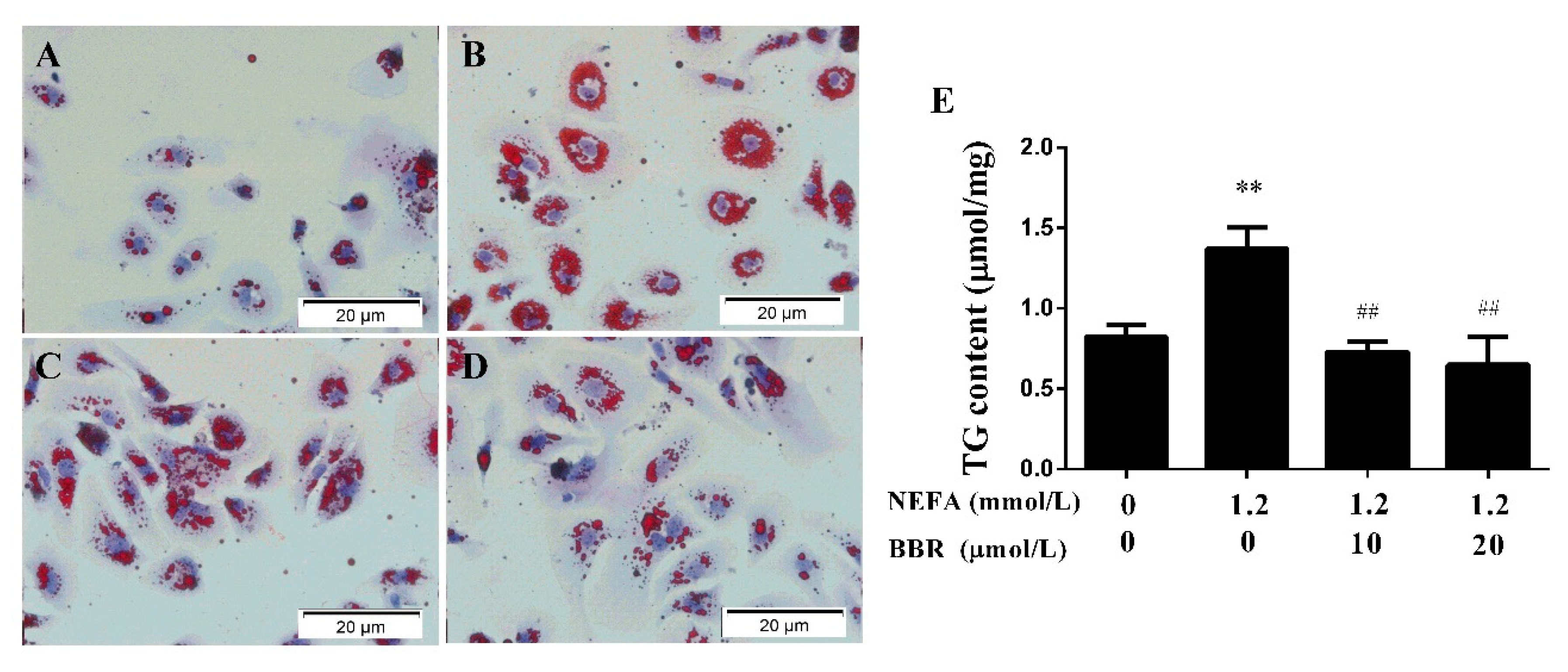
2.3. BBR Treatment Improved the NEFA-Induced Lipid Accumulation in Bovine Hepatocytes
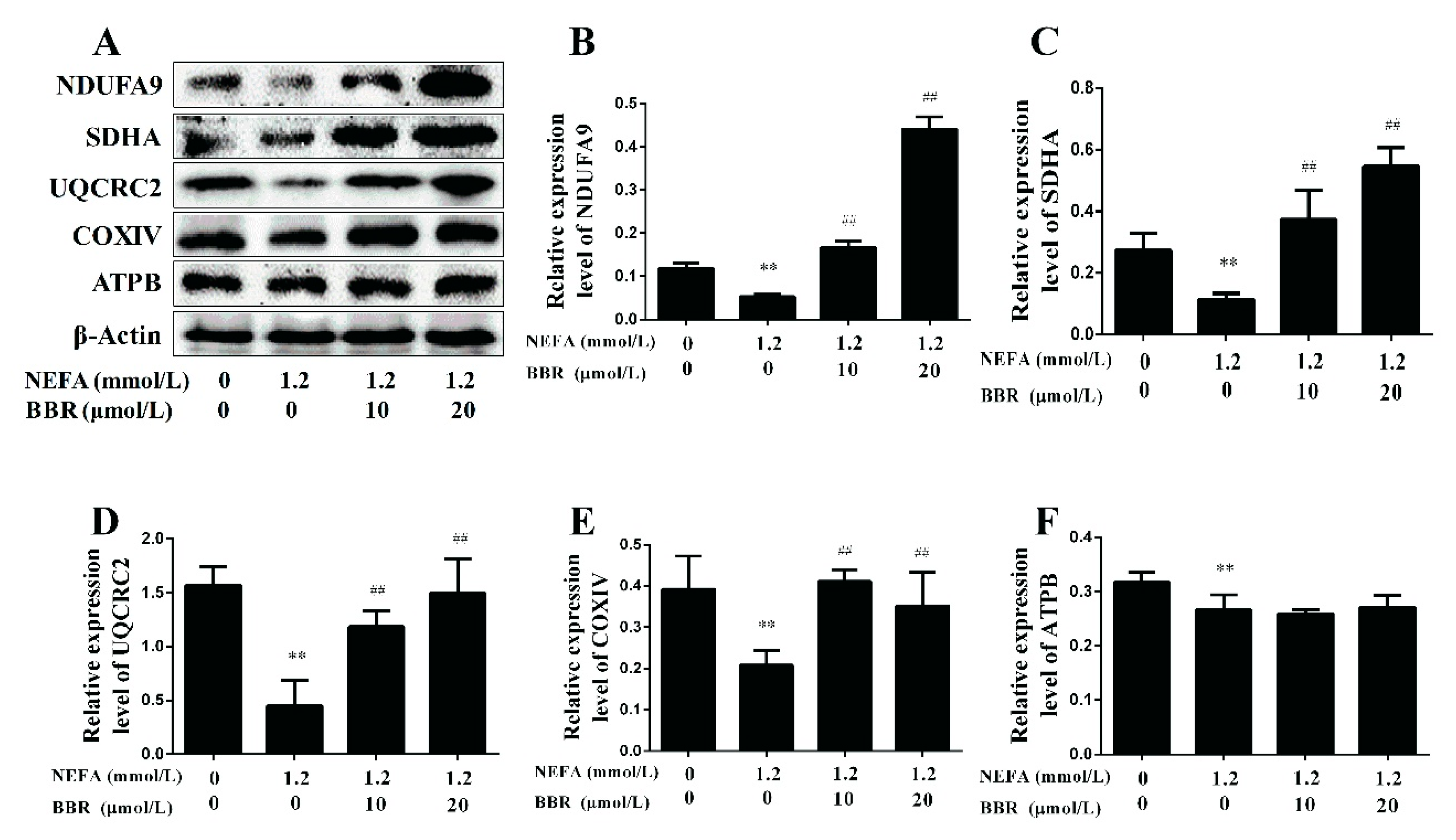
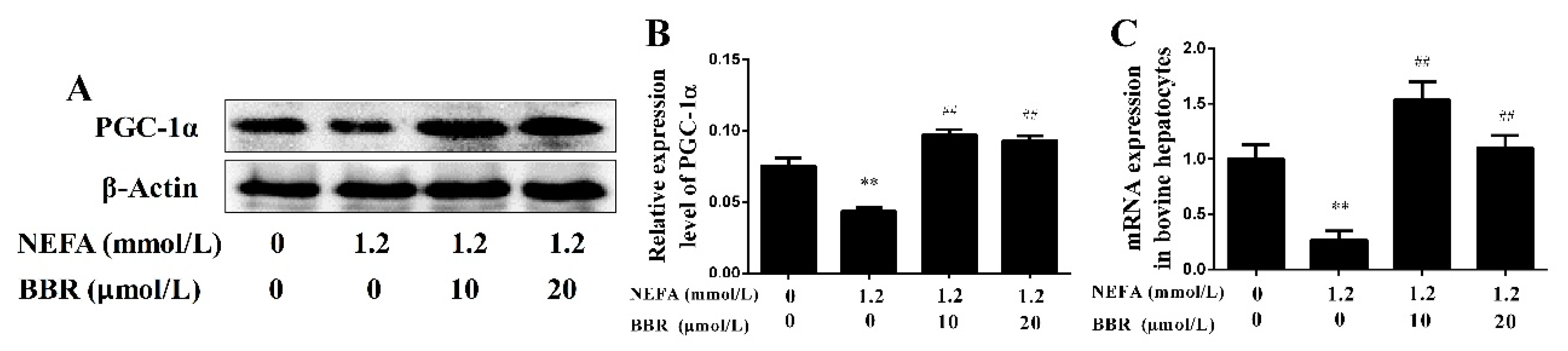
2.4. BBR Treatment Improved NEFA-Induced Mitochondrial Respiratory Chain Function and Insulin Signaling Damage by Increasing PGC-1α in Bovine Hepatocytes
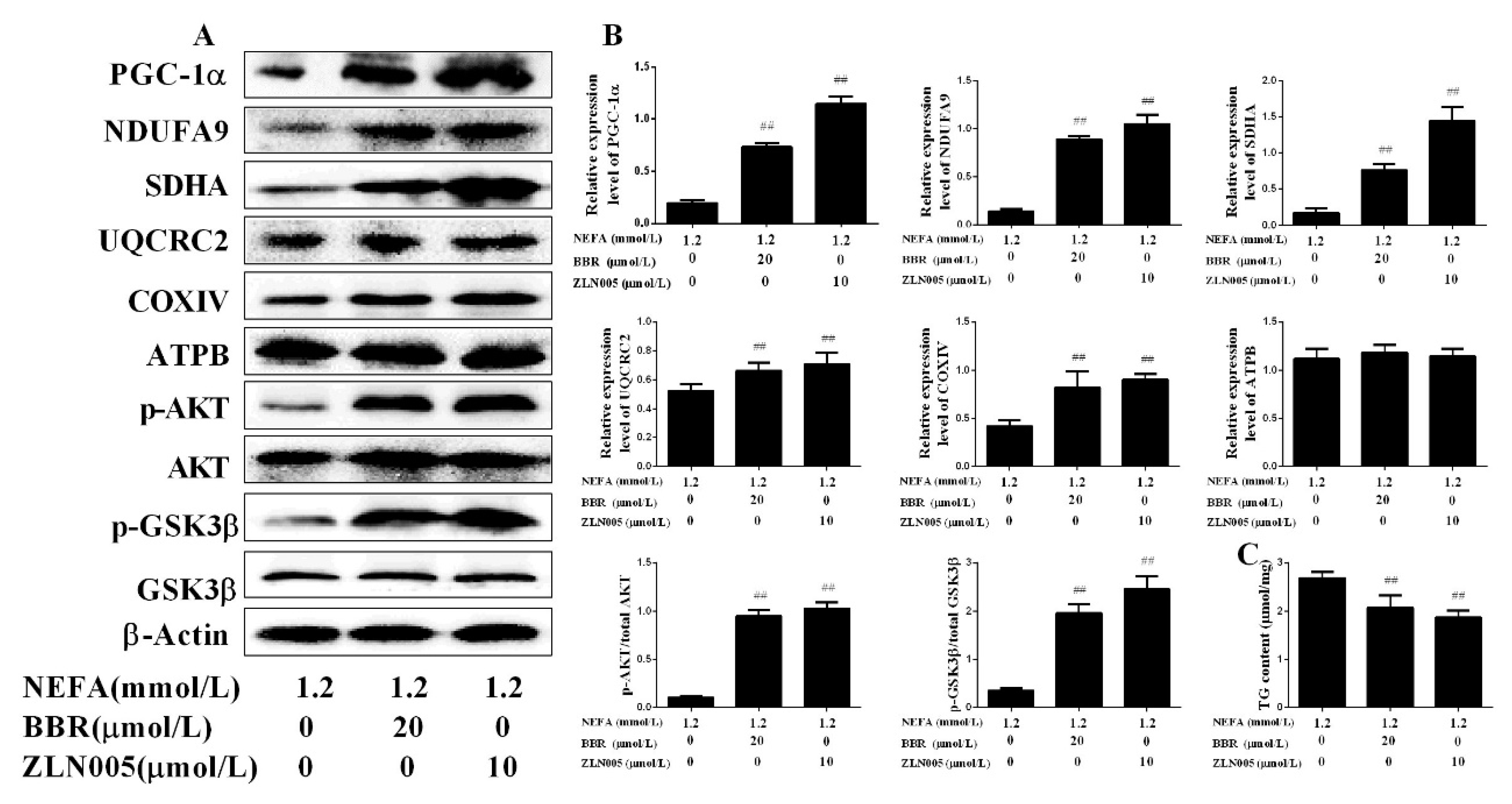
2.5. PGC-1α Mediated the Improvement Effect of BBR on NEFA-Induced Mitochondrial Respiratory Chain Function and Insulin Signaling Damage in Bovine Hepatocytes
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Determination of Blood Parameters
4.3. Bovine Hepatocyte Isolation and Culture
4.4. H&E Staining of Hepatic Tissue and Oil Red O Staining of Cow Hepatocytes
4.5. Determination of Hepatic and Cellular TG
4.6. RNA Extraction and Real-Time Quantitative PCR (qRT-PCR)
4.7. Western Blotting
4.8. Statistical Analysis
5. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Bobe, G.; Young, J.W.; Beitz, D.C. Invited review: Pathology, etiology, prevention, and treatment of fatty liver in dairy cows. J. Dairy Sci. 2004, 87, 3105–3124. [Google Scholar] [CrossRef]
- Jorritsma, R.; Jorritsma, H.; Schukken, Y.H.; Bartlett, P.C.; Wensing, T.; Wentink, G.H. Prevalence and indicators of post partum fatty infiltration of the liver in nine commercial dairy herds in the netherlands. Livestig. Prod. Sci. 2001, 68, 53–60. [Google Scholar] [CrossRef]
- Jorritsma, R.; Jorritsma, H.; Schukken, Y.H.; Wentink, G.H. Relationships between fatty liver and fertility and some periparturient diseases in commercial dutch dairy herds. Theriogenology 2000, 54, 1065–1074. [Google Scholar] [CrossRef]
- Oikawa, S.; Oetzel, G.R. Decreased insulin response in dairy cows following a four-day fast to induce hepatic lipidosis. J. Dairy Sci. 2006, 89, 2999–3005. [Google Scholar] [CrossRef]
- Xu, C.; Xu, Q.; Chen, Y.; Yang, W.; Cheng, X.; Yu, H.; Zhu, K.; Shen, T.; Zhang, Z. The relationship between fibroblast growth factor-21 and characteristic parameters related to energy balance in dairy cows. BMC Vet. Res. 2015, 11, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Li, X.; Li, Y.; Guan, Y.; Song, Y.; Yin, L.; Chen, H.; Lei, L.; Liu, J.; Li, X.; et al. Effects of nonesterified fatty acids on the synthesis and assembly of very low density lipoprotein in bovine hepatocytes in vitro. J. Dairy Sci. 2014, 97, 1328–1335. [Google Scholar] [CrossRef] [PubMed]
- Grummer, R.R. Nutritional and management strategies for the prevention of fatty liver in dairy cattle. Vet. J. 2008, 176, 10–20. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Huang, W.; Gu, J.; Du, X.; Lei, L.; Yuan, X.; Sun, G.; Wang, Z.; Li, X.; Liu, G. Srebp-1c overactivates ros-mediated hepatic NF-κB inflammatory pathway in dairy cows with fatty liver. Cell. Signal. 2015, 27, 2099–2109. [Google Scholar] [CrossRef] [PubMed]
- Miura, K.; Kodama, Y.; Inokuchi, S.; Schnabl, B.; Aoyama, T.; Ohnishi, H.; Olefsky, J.M.; Brenner, D.A.; Seki, E. Toll-like receptor 9 promotes steatohepatitis by induction of interleukin-1beta in mice. Gastroenterology 2010, 139, 323. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Xu, L.; Ye, J.; Li, D.; Wang, W.; Li, X.; Wu, L.; Wang, H.; Guan, F.; Li, P. Cidea promotes hepatic steatosis by sensing dietary fatty acids. Hepatology 2012, 56, 95–107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rector, R.S.; Thyfault, J.P.; Uptergrove, G.M.; Morris, E.M.; Naples, S.P.; Borengasser, S.J.; Mikus, C.R.; Laye, M.J.; Laughlin, M.H.; Booth, F.W. Mitochondrial dysfunction precedes insulin resistance and hepatic steatosis and contributes to the natural history of non-alcoholic fatty liver disease in an obese rodent model. J. Hepatol. 2010, 52, 727. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Feng, J.; Zhang, R.; Chen, J.; Han, D.; Li, X.; Yang, B.; Li, X.; Fan, M.; Li, C.; et al. Sirt1 activation by resveratrol alleviates cardiac dysfunction via mitochondrial regulation in diabetic cardiomyopathy mice. Oxid. Med. Cell. Longev. 2017, 2017, 4602715. [Google Scholar] [CrossRef] [PubMed]
- Choi, C.S.; Befroy, D.E.; Codella, R.; Kim, S.; Reznick, R.M.; Hwang, Y.J.; Liu, Z.X.; Lee, H.Y.; Distefano, A.; Samuel, V.T. Paradoxical effects of increased expression of PGC-1α on muscle mitochondrial function and insulin-stimulated muscle glucose metabolism. Proc. Natl. Acad. Sci. USA 2008, 105, 19926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, H.; Feng, L.; Xu, F.; Sun, Y.; Ma, Y.; Zhang, X.; Liu, H.; Xu, G.; Wu, X.; Shen, Y. Berberine inhibits palmitate-induced NLRP3 inflammasome activation by triggering autophagy in macrophages: A new mechanism linking berberine to insulin resistance improvement. Biomed. Pharmacother. 2017, 89, 864. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Li, Q.; Yu, B.; Yang, Y.; Jin, Z.; Duan, W.; Zhao, G.; Zhai, M.; Liu, L.; Yi, D.; et al. Berberine attenuates myocardial ischemia/reperfusion injury by reducing oxidative stress and inflammation response: Role of silent information regulator 1. Oxid. Med. Cell. Longev. 2016, 2016, 1689602. [Google Scholar] [CrossRef] [PubMed]
- Bova, S.; Padrini, R.; Goldman, W.F.; Berman, D.M.; Cargnelli, G. On the mechanism of vasodilating action of berberine: Possible role of inositol lipid signaling system. J. Pharmacol. Exp. Ther. 1992, 261, 318–323. [Google Scholar] [PubMed]
- Wang, N.; Xu, Q.; Tan, H.Y.; Hong, M.; Li, S.; Yuen, M.F.; Feng, Y. Berberine inhibition of fibrogenesis in a rat model of liver fibrosis and in hepatic stellate cells. Evid.-Based Complement. Altern. Med. ECAM 2016, 2016, 8762345. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Wang, G.; Yang, J.; Pan, X.; Yang, Z.; Zang, L. Berberine inhibits human hepatoma cell invasion without cytotoxicity in healthy hepatocytes. PLoS ONE 2011, 6, e21416. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, L.; Song, H.; Ji, G. Update on berberine in nonalcoholic fatty liver disease. Evid.-Based Complement. Altern. Med. ECAM 2013, 2013, 308134. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Yan, J.; Shen, Y.; Tang, K.; Yin, J.; Zhang, Y.; Yang, D.; Liang, H.; Ye, J.; Weng, J. Berberine improves glucose metabolism in diabetic rats by inhibition of hepatic gluconeogenesis. PLoS ONE 2011, 6, e16556. [Google Scholar] [CrossRef] [PubMed]
- Gomes, A.P.; Duarte, F.V.; Nunes, P.; Hubbard, B.P.; Teodoro, J.S.; Varela, A.T.; Jones, J.G.; Sinclair, D.A.; Palmeira, C.M.; Rolo, A.P. Berberine protects against high fat diet-induced dysfunction in muscle mitochondria by inducing sirt1-dependent mitochondrial biogenesis. Biochim. Biophys. Acta 2012, 1822, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Holtenius, P.; Holtenius, K. A model to estimate insulin sensitivity in dairy cows. Acta Vet. Scand. 2007, 49, 29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brusq, J.M.; Ancellin, N.; Grondin, P.; Guillard, R.; Martin, S.; Saintillan, Y.; Issandou, M. Inhibition of lipid synthesis through activation of amp kinase: An additional mechanism for the hypolipidemic effects of berberine. J. Lipid Res. 2006, 47, 1281–1288. [Google Scholar] [CrossRef] [PubMed]
- Bonnard, C.; Durand, A.; Peyrol, S.; Chanseaume, E.; Chauvin, M.A.; Morio, B.; Vidal, H.; Rieusset, J. Mitochondrial dysfunction results from oxidative stress in the skeletal muscle of diet-induced insulin-resistant mice. J. Clin. Investig. 2008, 118, 789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuo, J.J.; Chang, H.H.; Tsai, T.H.; Lee, T.Y. Positive effect of curcumin on inflammation and mitochondrial dysfunction in obese mice with liver steatosis. Int. J. Mol. Med. 2012, 30, 673. [Google Scholar] [CrossRef] [PubMed]
- Verbeek, J.; Lannoo, M.; Pirinen, E.; Ryu, D.; Spincemaille, P.; Elst, I.V.; Windmolders, P.; Thevissen, K.; Cammue, B.P.A.; Pelt, J.V. P245 roux-en-y gastric bypass attenuates hepatic mitochondrial dysfunction in mice with nonalcoholic steatohepatitis. J. Hepatol. 2014, 60, S147. [Google Scholar] [CrossRef]
- Wang, X.; West, J.A.; Murray, A.J.; Griffin, J.L. Comprehensive metabolic profiling of age-related mitochondrial dysfunction in the high-fat-fed ob/ob mouse heart. J. Proteome Res. 2015, 14, 2849–2862. [Google Scholar] [CrossRef] [PubMed]
- Michael, M.D.; Kulkarni, R.N.; Postic, C.; Previs, S.F.; Shulman, G.I.; Magnuson, M.A.; Kahn, C.R. Loss of insulin signaling in hepatocytes leads to severe insulin resistance and progressive hepatic dysfunction. Mol. Cell 2000, 6, 87–97. [Google Scholar] [CrossRef]
- Song, Y.; Li, X.; Li, Y.; Li, N.; Shi, X.; Ding, H.; Zhang, Y.; Li, X.; Liu, G.; Wang, Z. Non-esterified fatty acids activate the ros-p38-p53/nrf2 signaling pathway to induce bovine hepatocyte apoptosis in vitro. Apoptosis Int. J. Program. Cell Death 2014, 19, 984–997. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Li, D.; Deng, Q.; Li, Y.; Sun, G.; Yuan, X.; Song, Y.; Wang, Z.; Li, X.; Li, X.; et al. Nefas activate the oxidative stress-mediated NF-κB signaling pathway to induce inflammatory response in calf hepatocytes. J. Steroid Biochem. Mol. Biol. 2015, 145, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Teodoro, J.S.; Duarte, F.V.; Gomes, A.P.; Varela, A.T.; Peixoto, F.M.; Rolo, A.P.; Palmeira, C.M. Berberine reverts hepatic mitochondrial dysfunction in high-fat fed rats: A possible role for sirt3 activation. Mitochondrion 2013, 13, 637–646. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, Y.S.; Kim, W.S.; Kim, K.H.; Yoon, M.J.; Cho, H.J.; Shen, Y.; Ye, J.M.; Lee, C.H.; Oh, W.K.; Kim, C.T. Berberine, a natural plant product, activates AMP-activated protein kinase with beneficial metabolic effects in diabetic and insulin-resistant states. Diabetes 2006, 55, 2256. [Google Scholar] [CrossRef] [PubMed]
- Lou, T.; Zhang, Z.; Xi, Z.; Liu, K.; Li, L.; Liu, B.; Huang, F. Berberine inhibits inflammatory response and ameliorates insulin resistance in hepatocytes. Inflammation 2011, 34, 659–667. [Google Scholar] [CrossRef] [PubMed]
- Habib, J.; Niloofar, G.; Farahnaz, G.; Mehdi, B.; Ehsan, M.D.; Parviz, K. Effect of beberine on the survivin gene expression in peripheral blood mononuclear cell of chronic lymphocytic leukemia patients in vitro. Koomesh 2017, 19, 227–232. [Google Scholar]
- Song, Y.C.; Lee, Y.; Kim, H.M.; Hyun, M.Y.; Lim, Y.Y.; Song, K.Y.; Kim, B.J. Berberine regulates melanin synthesis by activating pi3k/akt, erk and gsk3beta in b16f10 melanoma cells. Int. J. Mol. Med. 2015, 35, 1011–1016. [Google Scholar] [CrossRef] [PubMed]
- Perseghin, G.; Caumo, A.; Caloni, M.; Testolin, G.; Luzi, L. Incorporation of the fasting plasma ffa concentration into quicki improves its association with insulin sensitivity in nonobese individuals. J. Clin. Endocrinol. Metab. 2001, 86, 4776–4781. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Shi, Z.; Peng, Z.; Zhao, C.; Zhang, Y.; Wang, Z.; Li, X.; Liu, G.; Li, X. Acetoacetate induces hepatocytes apoptosis by the ROS-mediated mapks pathway in ketotic cows. J. Cell. Physiol. 2017, 232, 3296–3308. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Li, N.; Gu, J.; Fu, S.; Peng, Z.; Zhao, C.; Zhang, Y.; Li, X.; Wang, Z.; Li, X.; et al. Beta-hydroxybutyrate induces bovine hepatocyte apoptosis via an ros-p38 signaling pathway. J. Dairy Sci. 2016, 99, 9184–9198. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Yuan, X.; Chen, L.; Wang, T.; Wang, Z.; Sun, G.; Li, X.; Li, X.; Liu, G. Histamine induces bovine rumen epithelial cell inflammatory response via NF-κB pathway. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2017, 42, 1109–1119. [Google Scholar] [CrossRef] [PubMed]




| Blood Biochemical Index | Healthy Cows | Cows with Fatty Liver | p Value |
|---|---|---|---|
| Body weight (kg) | 536.8 ± 19.1 | 541.3 ± 18.2 | 0.779 |
| INSULIN (mU/L) | 1.45 ± 0.12 | 1.96 ± 0.16 * | 0.011 |
| GLU (mmol/L) | 3.26 ± 0.44 | 2.09 ± 0.07 * | 0.041 |
| ALT (U/L) | 14.79 ± 1.42 | 23.80 ± 1.29 ** | 0.001 |
| AST (U/L) | 80.79 ± 15.67 | 170.97 ± 9.25 ** | 0.001 |
| AST/ALT | 5.44 ± 0.68 | 7.18 ± 0.11 * | 0.044 |
| ALP (U/L) | 67.5 ± 3.31 | 44.5 ± 4.35 ** | 0.002 |
| TBIL (μmol/L) | 2.37 ± 0.10 | 6.09 ± 0.78 ** | 0.001 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, Z.; Li, X.-B.; Peng, Z.-C.; Fu, S.-P.; Zhao, C.-X.; Du, X.-L.; Fang, Z.-Y.; Wang, Z.; Liu, G.-W.; Li, X.-W. Berberine Protects against NEFA-Induced Impairment of Mitochondrial Respiratory Chain Function and Insulin Signaling in Bovine Hepatocytes. Int. J. Mol. Sci. 2018, 19, 1691. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19061691
Shi Z, Li X-B, Peng Z-C, Fu S-P, Zhao C-X, Du X-L, Fang Z-Y, Wang Z, Liu G-W, Li X-W. Berberine Protects against NEFA-Induced Impairment of Mitochondrial Respiratory Chain Function and Insulin Signaling in Bovine Hepatocytes. International Journal of Molecular Sciences. 2018; 19(6):1691. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19061691
Chicago/Turabian StyleShi, Zhen, Xiao-Bing Li, Zhi-Cheng Peng, Shou-Peng Fu, Chen-Xu Zhao, Xi-Liang Du, Zhi-Yuan Fang, Zhe Wang, Guo-Wen Liu, and Xin-Wei Li. 2018. "Berberine Protects against NEFA-Induced Impairment of Mitochondrial Respiratory Chain Function and Insulin Signaling in Bovine Hepatocytes" International Journal of Molecular Sciences 19, no. 6: 1691. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19061691




