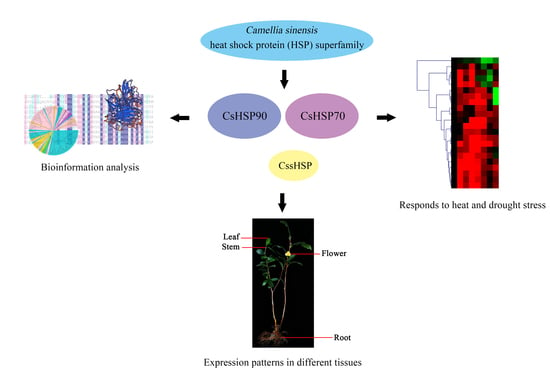
Genome-Wide Identification, Classification and Expression Analysis of the HSP Gene Superfamily in Tea Plant (Camellia sinensis)
Abstract
:1. Introduction
2. Results
2.1. Identification and Characterization of the CsHSP Gene Superfamily in Tea Plant
2.2. Phylogenetic Analysis of CsHSPs
2.3. Gene Structures of the CsHSP Gene Superfamily
2.4. Conserved Protein Motif Analysis of CsHSPs
2.5. Protein Interaction Network of CsHSPs
2.6. Expression Patterns of CsHSP Genes in Different Tissues of Tea Plant
2.7. Expression Profiles of CsHSP Genes in Response to Heat and Drought Stress
3. Discussion
4. Materials and Methods
4.1. Plant Materials, Growth Conditions, and Stress Treatments
4.2. Identification of the CsHSP Gene Superfamily in Tea Plant
4.3. Sequence Analysis and Phylogenetic Tree Construction
4.4. Conserved Motif Analysis and Protein Interaction Network Prediction of CsHSPs
4.5. RNA Isolation and Expression Analysis of CsHSP Genes
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Mittler, R. Abiotic stress, the field environment and stress combination. Trends Plant Sci. 2006, 11, 15–19. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R.; Finka, A.; Goloubinoff, P. How do plants feel the heat? Trends Biochem. Sci. 2012, 37, 118–125. [Google Scholar] [CrossRef] [PubMed]
- Montero-Barrientos, M.; Hermosa, R.; Nicolás, C.; Cardoza, R.E.; Gutiérrez, S.; Monte, E. Overexpression of a Trichoderma HSP70 gene increases fungal resistance to heat and other abiotic stresses. Fungal Genet. Biol. 2008, 45, 1506–1513. [Google Scholar] [CrossRef] [PubMed]
- Balfagóna, D.; Zandalinasa, S.I.; Baliñob, P.; Muriachb, M.; Gómez-Cadenas, A. Involvement of ascorbate peroxidase and heat shock proteins on citrus tolerance to combined conditions of drought and high temperatures. Plant Physiol. Biochem. 2018, 127, 194–199. [Google Scholar] [CrossRef] [PubMed]
- Park, C.; Seo, Y. Heat Shock Proteins: A Review of the molecular chaperones for plant immunity. Plant Pathol. J. 2015, 31, 323–333. [Google Scholar] [CrossRef] [PubMed]
- Vierling, E. The Roles of heat shock proteins in plants. Annu. Rev. Plant Physiol. 1991, 42, 579–620. [Google Scholar] [CrossRef]
- Sung, D.Y.; Kaplan, F.; Lee, K.J.; Guy, C.L. Acquired tolerance to temperature extremes. Trends Plant Sci. 2003, 8, 179–187. [Google Scholar] [CrossRef] [Green Version]
- Charng, Y.; Liu, H.; Liu, N.; Hsu, F.; Ko, S. Arabidopsis Hsa32, a novel heat shock protein, is essential for acquired thermotolerance during long recovery after acclimation. Plant Physiol. 2014, 140, 1297–1305. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, N.K.; Kim, Y.K.; Grover, A. Rice sHsp genes: Genomic organization and expression profiling under stress and development. BMC Genom. 2009, 10, 393. [Google Scholar] [CrossRef] [PubMed]
- Waters, E.R. The evolution, function, structure, and expression of the plant sHSPs. J. Exp. Bot. 2013, 64, 391–403. [Google Scholar] [CrossRef] [PubMed]
- Timperio, A.M.; Egidi, M.G.; Zolla, L. Proteomics applied on plant abiotic stresses: Role of heat shock proteins (HSP). J. Proteom. 2008, 71, 391–411. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, J.; Liu, B.; Zhang, L.; Chen, J.; Lu, M. Genome-wide analysis of the populus Hsp90 gene family reveals differential expression patterns, localization, and heat stress responses. BMC Genom. 2013, 14, 532. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Vinocur, B.; Shoseyov, O.; Altman, A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 2004, 9, 244–252. [Google Scholar] [CrossRef] [PubMed]
- Lin, B.; Wang, J.; Liu, H.; Chen, R.; Meyer, Y.; Barakat, A.; Delseny, M. Genomic analysis of the Hsp70 superfamily in Arabidopsis thaliana. Cell Stress Chaperon. 2001, 6, 201–208. [Google Scholar] [CrossRef]
- Scharf, K.D.; Siddique, M.; Vierling, E. The expanding family of Arabidopsis thaliana small heat stress proteins and a new family of proteins containing a-crystallin domains (acd proteins). Cell Stress Chaperon. 2001, 6, 225–237. [Google Scholar] [CrossRef]
- Krishna, P.; Gloor, G. The HSP90 family of proteins in Arabidopsis thaliana. Cell Stress Chaperon. 2001, 6, 238–246. [Google Scholar] [CrossRef]
- Dimitra Milioni, D.; Hatzopoulos, P. Genomic organization of hsp90 gene family in Arabidopsis. Plant Mol. Biol. 1997, 35, 955–961. [Google Scholar] [CrossRef]
- Li, Z.; Long, R.; Zhang, T.; Wang, Z.; Zhang, F.; Yang, Q.; Kang, J.; Sun, Y. Molecular cloning and functional analysis of the drought tolerance gene MsHSP70 from alfalfa (Medicago sativa L.). J. Plant Res. 2017, 130, 387–396. [Google Scholar] [CrossRef] [PubMed]
- Masand, S.; Yadav, S.K. Overexpression of MuHSP70 gene from Macrotyloma uniflorum confers multiple abiotic stress tolerance in transgenic Arabidopsis thaliana. Mol. Biol. Rep. 2016, 43, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Aviezer-Hagai, K.; Skovorodnikova, J.; Galigniana, M.; Farchi-Pisanty, O.; Maayan, E.; Bocovza, S.; Efrat, Y.; von Koskull-Döring, P.; Ohad, N.; Breiman, A. Arabidopsis immunophilins ROF1 (AtFKBP62) and ROF2 (AtFKBP65) exhibit tissue specificity, are heat-stress induced, and bind HSP90. Plant Mol. Biol. 2007, 63, 237–255. [Google Scholar] [CrossRef] [PubMed]
- Meiri, D.; Breiman, A. Arabidopsis ROF1 (FKBP62) modulates thermotolerance by interacting with HSP90.1 and affecting the accumulation of HsfA2-regulated sHSPs. Plant J. 2009, 59, 387–399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Hartl, F.; Bracher, A. Structure and function of Hip, an attenuator of the Hsp70 chaperone cycle. Nat. Struct. Mol. Biol. 2013, 20, 929–935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernández-Bautista, N.; Fernández-Calvino, L.; Muñoz, A.; Castellano, M.M. HOP3, a member of the HOP family in Arabidopsis, interacts with BiP and plays a major role in the ER stress response. Plant Cell Environ. 2017, 40, 1341. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.H.; Alam, I.; Kim, Y.G.; Sharmin, S.A.; Lee, K.W.; Lee, S.H.; Lee, B.H. Overexpression of a chloroplast-localized small heat shock protein OsHSP26 confers enhanced tolerance against oxidative and heat stresses in tall fescue. Biotechnol. Lett. 2012, 34, 371–377. [Google Scholar] [CrossRef] [PubMed]
- Merino, I.; Contreras, A.; Jing, Z.; Gallardo, F.; Francisco, M.; Cánovas, F.M.; Gómez, L. Plantation forestry under global warming: Hybrid poplars with improved thermotolerance provide new insights on the in vivo function of small heat shock protein chaperones. Plant Physiol. 2014, 164, 978–991. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Yan, H.; Wang, Y.; Feng, L.; Chen, Z.; Xiang, Y. Genome duplication and evolution of heat shock transcription factor (HSF) gene family in four model angiosperms. J. Plant Growth Regul. 2016, 35, 903–920. [Google Scholar] [CrossRef]
- Jacob, P.; Hirt, H.; Bendahmane, A. The heat-shock protein/chaperone network and multiple stress resistance. Plant Biotechnol. J. 2017, 15, 405–414. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, Y.; Chen, J.; Xie, W.; Wang, L.; Zhang, Q. Comprehensive sequence and expression profile analysis of Hsp20 gene family in rice. Plant Mol. Biol. 2009, 70, 341–357. [Google Scholar] [CrossRef] [PubMed]
- Lopes-Caitar, V.S.; de Carvalho, M.C.; Darben, M.; Kuwahara, M.K.; Nepomuceno, A.L.; Dias, W.P.; Abdelnoor, R.V.; Marcelino-Guimarães, F.C. Genome-wide analysis of the Hsp20 gene family in soybean: Comprehensive sequence, genomic organization and expression profile analysis under abiotic and biotic stresses. BMC Genom. 2013, 14, 577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yer, E.N.; Baloglu, M.C.; Ziplar, U.T.; Ayan, S.; Unver, T. Drought-responsive Hsp70 gene analysis in populus at genome-wide level. Plant Mol. Biol. Rep. 2016, 34, 483–500. [Google Scholar] [CrossRef]
- Banilas, G.; Korkas, E.; Engezos, V.; Nisiotou, A.A.; Hatzopouos, P. Genome-wide analysis of the heat shock protein 90 gene family in grapevine (Vitis vinifera L.). Aust. J. Grape Wine Res. 2012, 18, 29–38. [Google Scholar] [CrossRef]
- Song, D.; Jung, Y.; Chay, K.; Chung, M.A.; Lee, K.H.; Yang, S.Y.; Shin, B.A.; Ahn, B.W. Effect of drinking green tea on age-associated accumulation of maillard-type fluorescence and carbonyl groups in rat aortic and skin collagen. Arch. Biochem. Biophys. 2002, 397, 424–429. [Google Scholar] [CrossRef] [PubMed]
- Bordonia, A.; Hrelia, S.; Angelonia, C.; Giordanob, E.; Guarnierib, C.; Caldarerab, C.M.; Biagia, P.L. Green tea protection of hypoxia/reoxygenation injury in cultured cardiac cells. J. Nutr. Biochem. 2002, 13, 103–111. [Google Scholar] [CrossRef]
- Wang, W.; Xin, H.; Wang, M.; Ma, Q.; Wang, L.; Kaleri, N.A.; Wang, Y.; Li, X. Transcriptomic analysis of Camellia sinensis reveals the molecular mechanisms of drought-stress-induced decreases in leaf quality. Front. Plant Sci. 2016, 7, 385. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zou, Z.; Li, Q.; Xin, H.; Zhu, X.; Chen, X.; Li, X. Heterologous expression of three Camellia sinensis small heat shock protein genes confers temperature stress tolerance in yeast and Arabidopsis thaliana. Plant Cell Rep. 2017, 36, 1125–1135. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zou, Z.; Li, Q.; Sun, K.; Chen, X.; Li, X. The CsHSP17.2 molecular chaperone is essential for thermotolerance in Camellia sinensis. Sci. Rep. 2016, 7, 1237. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Yang, H.; Wang, S.; Zhao, J.; Liu, C.; Gao, L.; Xia, E.; Lu, Y.; Tai, Y.; She, G.; et al. Draft genome sequence of Camellia sinensis var. sinensis provides insights into the evolution of the tea genome and tea quality. Proc. Natl. Acad. Sci. USA 2018, 115, E4151–E4158. [Google Scholar] [PubMed]
- Siddique, M.; Gernhard, S.; von Koskull-Döring, P.; Vierling, E.; Scharf, K.D. The plant sHSP superfamily: Five new members in Arabidopsis thaliana with unexpected properties. Cell Stress Chaperon. 2008, 13, 183–197. [Google Scholar] [CrossRef] [PubMed]
- Muthusamy, S.D.; Dalala, M.; Chinnusamyc, V.; Bansal, K.C. Genome-wide identification and analysis of biotic and abiotic stress regulation of small heat shock protein (HSP20) family genes in bread wheat. J. Plant Physiol. 2017, 211, 100–113. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Li, X.; Liu, Z.; Li, H.; Wang, Y.; Zhuang, J. Transcriptome-wide identification of Camellia sinensis WRKY transcription factors in response to temperature stress. Mol. Genet. Genom. 2016, 291, 255–269. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, C.; Ban, F.; Zhu, D.; Liu, S.; Wang, X. Genome-wide identification and characterization of HSP gene superfamily in whitefly (Bemisia tabaci) and expression profiling analysis under temperature stress. Insect Sci. 2017. [Google Scholar] [CrossRef] [PubMed]
- Dragovic, Z.; Broadley, S.A.; Shomura, Y.; Bracher, A.; Hartl, F.U. Molecular chaperones of the Hsp110 family act as nucleotide exchange factors of Hsp70s. EMBO J. 2006, 25, 2519–2528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, M.; Liu, J.; Lu, J.; Zhai, Y.; Wang, H.; Gong, Z.; Wang, S.; Lu, M. Genome-wide analysis of the CaHsp20 gene family in pepper: Comprehensive sequence and expression profile analysis under heat stress. Front. Plant Sci. 2015, 6, 806. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Cheng, Y.; Feng, K.; Ruan, M.; Ye, Q.; Wang, R.; Li, Z.; Zhou, G.; Yao, Z.; Yang, Y.; et al. Genome-wide identification and expression profiling of tomato Hsp20 gene family in response to biotic and abiotic stresses. Front. Plant Sci. 2016, 7, 1215. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Zhang, A.; Chen, J.; He, X.; Xu, B.; Xie, G.; Miao, Z.; Zhang, X.; Huang, L. Genome-wide analysis of the PvHsp20 family in switchgrass: Motif, genomic organization, and identification of stress or developmental-related Hsp20s. Front. Plant Sci. 2017, 8, 1024. [Google Scholar] [CrossRef] [PubMed]
- Xua, G.; Guo, C.; Shan, H.; Kong, H. Divergence of duplicate genes in exon-intron structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1187–1192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wehmeyer, N.; Vierling, E. The expression of small heat shock proteins in seeds responds to discrete developmental signals and suggests a general protective role in desiccation tolerance. Plant Physiol. 2000, 122, 1099–1108. [Google Scholar] [CrossRef] [PubMed]
- Yabe, N.; Takahashi, T.; Komeda, T. Analysis of tissue-specific expression of Arabidopsis thaliana HSP90-family gene HSP81. Plant Cell Physiol. 1994, 35, 1207. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Li, L.; Ye, T.; Chen, R.; Gao, X.; Xu, Z. Molecular characterization, expression pattern and function analysis of the OsHSP90 family in rice. Biotechnol. Biotechnol. Equip. 2016, 30, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Mertz-Henning, L.M.; Pegoraro, C.; Maia, L.C.; Venske, E.; Rombaldi, C.V.; de Oliveira, C.A. Expression profile of rice Hsp genes under anoxic stress. Genet. Mol. Res. 2016, 15. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, N.K.; Kundnani, P.; Grover, A. Functional analysis of Hsp70 superfamily proteins of rice (Oryza sativa). Cell Stress Chaperon. 2013, 18, 427–437. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, B.; Li, J.; Zhang, L.; Wang, Y.; Zheng, H.; Lu, M.; Chen, J. Hsf and Hsp gene families in populus: Genome-wide identification, organization and correlated expression during development and in stress responses. BMC Genom. 2015, 16, 181. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhao, H.; Dong, Q.; Zhang, Y.; Wang, Y.; Li, H.; Xing, G.; Li, Q.; Dong, Y. Genome-wide analysis and expression profiling under heat and drought treatments of HSP70 gene family in soybean (Glycine max L.). Front. Plant Sci. 2015, 6, 773. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Bautista, N.; Fernández-Calvino, L.; Muñoz, A.; Toribio, R.; Mock, H.P.; Castellano, M.M. HOP family plays a major role in long term acquired thermotolerance in Arabidopsis. Plant Cell Environ. 2018, 41, 1852–1869. [Google Scholar] [CrossRef] [PubMed]
- Cha, J.Y.; Baek, D.; Lee, K.W.; Cheong, M.S.; Hong, S.; Son, D. Identification of heat shock-inducible transcripts and Hop (Hsp-organizing protein)-interacting proteins in orchardgrass (Dactylis glomerata L.). Acta Physiol. Plant. 2018, 40, 132. [Google Scholar] [CrossRef]
- Makhnevych, T.; Houry, W.A. The control of spindle length by Hsp70 and Hsp110 molecular chaperones. FEBS Lett. 2013, 587, 1067–1072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yam, A.Y.W.; Albanèse, V.; Lin, H.T.J.; Frydman, J. HSP110 cooperates with different cytosolic HSP70 systems in a pathway for de novo folding. J. Biol. Chem. 2005, 280, 41252. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.J.; Vierling, E. A small heat shock protein cooperates with heat shock protein 70 systems to reactivate a heat-denatured protein. Plant Physiol. 2002, 122, 189–197. [Google Scholar] [CrossRef]
- Mogk, A.; Deuerling, E.; Vorderwülbecke, S.; Vierling, E.; Bukau, B. Small heat shock proteins, ClpB and the DnaK system form a functional triade in reversing protein aggregation. Mol. Microbiol. 2003, 50, 585–595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Charng, Y. Common and distinct functions of Arabidopsis cass A1 and A2 heat shock factors in diverse abiotic stress responses and development. Plant Physiol. 2013, 163, 276–290. [Google Scholar] [CrossRef] [PubMed]
- Hahn, A.; Bublak, D.; Schleiff, E.; Scharf, K.D. Crosstalk between HSP90 and HSP70 chaperones and heat stress transcription factors in tomato. Plant Cell 2011, 23, 741–755. [Google Scholar] [CrossRef] [PubMed]
- Xia, E.; Zhang, H.; Sheng, J.; Li, K.; Zhang, Q.; Kim, C.; Zhang, Y.; Liu, Y.; Zhu, T.; Li, W.; et al. The tea tree genome provides insights into tea flavor and independent evolution ofcaffeine biosynthesis. Mol. Plant. 2017, 6, 866–877. [Google Scholar] [CrossRef] [PubMed]
- Hao, X.; Horvath, D.P.; Chao, W.; Yang, Y.; Wang, X.; Xiao, B. Identification and evaluation of reliable reference genes for quantitative real-time PCR analysis in tea plant (Camellia sinensis (L.) O. Kuntze). Int. J. Mol. Sci. 2014, 15, 22155–22172. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−△△Ct method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]








| Name | Unigene ID | Animo Acids | MW (kDa) | pI | GRAVY | Instability Index | Aliphatic Index | Subcellular Localization |
|---|---|---|---|---|---|---|---|---|
| CsHSP90-1 | TEA014732.1 | 790 | 89.90 | 5.20 | −0.58 | 34.37 | 77.62 | mito: 7, chlo: 4, nucl: 2, cysk: 1 |
| CsHSP90-2 | TEA027510.1 | 823 | 94.33 | 4.84 | −0.75 | 37.01 | 79.62 | E.R.: 14 |
| CsHSP90-3 | TEA017408.1 | 810 | 92.31 | 5.01 | −0.60 | 43.77 | 80.26 | chlo: 11.5, chlo_mito: 7.5, mito: 2.5 |
| CsHSP90-4 | TEA027510.1 | 701 | 80.44 | 5.11 | −0.60 | 35.08 | 84.95 | cyto: 7, nucl: 2, plas: 2, mito: 1, vacu: 1, golg: 1 |
| CsHSP90-5 | TEA008112.1 | 699 | 79.99 | 4.99 | −0.56 | 36.68 | 84.23 | cyto: 8, plas: 2, E.R.: 2, mito: 1, golg: 1 |
| CsHSP90-6 | TEA007238.1 | 707 | 81.14 | 5.04 | −0.59 | 42.75 | 82.26 | nucl: 6, cyto: 5, chlo: 2, cysk: 1 |
| CsHSP90-7 | TEA027790.1 | 643 | 73.68 | 5.11 | −0.50 | 39.33 | 85.99 | cyto: 9, nucl: 3, plas: 1, golg: 1 |
| CsHSP70-1 | TEA005970.1 | 648 | 71.03 | 5.16 | −0.42 | 34.56 | 81.87 | cyto: 7, cysk: 6, chlo: 1 |
| CsHSP70-2 | TEA029156.1 | 846 | 93.33 | 5.21 | −0.42 | 44.20 | 80.18 | cyto: 8, chlo: 2, nucl: 2, cysk: 2 |
| CsHSP70-3 | TEA023352.1 | 892 | 98.89 | 5.22 | −0.46 | 41.33 | 83.55 | E.R.: 14 |
| CsHSP70-4 | TEA003665.1 | 709 | 75.51 | 5.38 | −0.29 | 33.70 | 85.51 | chlo: 14 |
| CsHSP70-5 | TEA016684.1 | 569 | 61.81 | 5.54 | 0.09 | 45.27 | 100.39 | cyto: 11, chlo: 2, nucl: 1 |
| CsHSP70-6 | TEA022542.1 | 679 | 73.09 | 6.02 | −0.33 | 41.82 | 86.07 | mito: 12, chlo: 2 |
| CsHSP70-7 | TEA023884.1 | 669 | 73.73 | 5.03 | −0.45 | 31.40 | 86.43 | E.R.: 14 |
| CsHSP70-8 | CL5657.Contig2_All▲ | 651 | 71.19 | 5.20 | −0.45 | 34.66 | 80.06 | cyto: 12, chlo: 1, nucl: 1 |
| CsHSP70-9 | TEA025820.1 | 677 | 75.12 | 5.54 | −0.27 | 34.56 | 87.70 | plas: 11, nucl: 1, vacu: 1, E.R.: 1 |
| CsHSP70-10 | TEA024518.1 | 744 | 82.42 | 5.79 | −0.43 | 32.96 | 82.34 | cyto: 8, cysk: 5, chlo: 1 |
| CsHSP70-11 | TEA023111.1 | 669 | 74.02 | 5.14 | −0.42 | 31.86 | 87.43 | plas: 11.5, golg_plas: 7, golg: 1.5, E.R.: 1 |
| CsHSP70-12 | CSA029398.1◆ | 519 | 57.32 | 6.15 | −0.31 | 39.76 | 88.86 | cysk: 11, chlo: 2, cyto: 1 |
| CsHSP70-13 | TEA014652.1 | 614 | 67.30 | 5.43 | −0.37 | 31.36 | 85.94 | chlo: 9, nucl: 1, cyto: 1, extr: 1, vacu: 1, pero: 1 |
| CsHSP70-14 | TEA024522.1 | 574 | 63.62 | 7.99 | −0.43 | 37.81 | 85.45 | cyto: 11, chlo: 2, nucl: 1 |
| CsHSP70-15 | TEA029776.1 | 627 | 69.61 | 5.07 | 0.01 | 36.23 | 106.67 | cyto: 9, chlo: 2, nucl: 2, cysk: 1 |
| CsHSP70-16 | TEA029256.1 | 449 | 49.87 | 5.32 | −0.52 | 40.99 | 79.73 | extr: 6, cyto: 3, chlo: 2, vacu: 1, E.R.: 1, mito_plas: 1 |
| CsHSP70-17 | TEA024511.1 | 407 | 45.46 | 9.04 | −0.38 | 32.41 | 87.00 | chlo: 11, mito: 1, extr: 1, E.R.: 1 |
| CsHSP70-18 | TEA017380.1 | 767 | 85.85 | 5.60 | −0.41 | 43.64 | 83.42 | nucl: 6, chlo: 3, cyto: 2, cysk: 2, vacu: 1 |
| CssHSP-1 | TEA023567.1 | 136 | 15.46 | 5.60 | −0.18 | 58.91 | 88.16 | cyto: 10, chlo: 1, nucl: 1, extr: 1, golg: 1 |
| CssHSP-2 | TEA025531.1 | 155 | 17.42 | 5.13 | −0.57 | 52.60 | 80.52 | cyto: 13, chlo: 1 |
| CssHSP-3 | TEA030322.1 | 199 | 22.45 | 5.99 | −0.56 | 52.92 | 88.14 | mito: 11, chlo: 3 |
| CssHSP-4 | TEA028966.1 | 243 | 27.40 | 8.29 | −0.71 | 43.78 | 62.55 | chlo: 13, nucl: 1 |
| CssHSP-5 | TEA031630.1 | 255 | 28.08 | 8.76 | −0.43 | 60.82 | 69.25 | chlo: 10, nucl: 2, extr: 2 |
| CssHSP-6 | TEA004366.1 | 159 | 18.25 | 5.68 | −0.70 | 62.21 | 71.70 | cyto: 14 |
| CssHSP-7 | TEA031692.1 | 153 | 17.47 | 5.53 | −0.61 | 54.38 | 73.20 | cyto: 14 |
| CssHSP-8 | TEA033542.1 | 223 | 25.19 | 5.83 | −0.53 | 46.89 | 81.26 | chlo: 10, mito: 2, vacu: 2 |
| CssHSP-9 | TEA015431.1 | 150 | 17.25 | 5.56 | −0.71 | 44.10 | 68.20 | cyto: 13, extr: 1 |
| CssHSP-10 | CL9235.Contig2_All▲ | 144 | 16.08 | 6.76 | −0.49 | 41.09 | 83.12 | pero: 8, nucl: 2, cyto: 2, chlo: 1, golg: 1 |
| CssHSP-11 | CSA020588.1◆ | 159 | 18.34 | 9.81 | −0.56 | 45.22 | 83.96 | cyto: 6, mito: 5, nucl: 3 |
| CssHSP-12 | CSA025700.1◆ | 196 | 21.66 | 5.30 | −0.71 | 52.67 | 68.67 | cyto: 8, nucl: 6 |
| CssHSP-13 | TEA032865.1 | 149 | 17.05 | 8.83 | −0.72 | 39.98 | 69.93 | cyto: 14 |
| CssHSP-14 | TEA015431.1 | 149 | 17.11 | 5.38 | −0.65 | 44.27 | 68.66 | cyto: 7, nucl: 3, extr: 2, chlo: 1, plas: 1 |
| CssHSP-15 | TEA010603.1 | 198 | 22.77 | 4.83 | −0.58 | 46.14 | 69.34 | chlo: 5, nucl: 5, cyto: 3, plas: 1 |
| CssHSP-16 | TEA021291.1 | 195 | 22.15 | 5.29 | −0.59 | 43.43 | 70.36 | nucl: 5, cyto: 5, chlo: 3, plas: 1 |
| CssHSP-17 | TEA033850.1 | 164 | 18.08 | 5.78 | −0.37 | 39.30 | 83.84 | cyto: 8, nucl: 5, chlo: 1 |
| CssHSP-18 | TEA005909.1 | 205 | 22.91 | 9.52 | −0.41 | 32.13 | 84.73 | cyto: 3, golg: 3, plas: 2.5, E.R.plas: 2.5, mito: 2, E.R.: 1.5, chlo: 1, vacu: 1 |
| CssHSP-19 | TEA005935.1 | 199 | 20.95 | 8.93 | −0.28 | 47.34 | 76.58 | E.R.: 3.5, golg: 3, cyto: 2.5, E.R.plas: 2.5, mito: 2, cyto_pero: 2, chlo: 1, vacu:1 |
| CssHSP-20 | TEA004364.1 | 249 | 27.72 | 9.49 | −0.43 | 48.28 | 74.90 | mito: 9, nucl: 3, chlo: 2 |
| CssHSP-21 | TEA028346.1 | 140 | 16.25 | 6.77 | −0.88 | 42.55 | 74.43 | cyto: 5, extr: 4, nucl: 2, golg: 2, plas: 1 |
| CssHSP-22 | TEA017741.1 | 188 | 21.78 | 6.47 | −0.49 | 49.13 | 92.82 | chlo: 7, mito: 3, cyto: 2, extr: 1, E.R.: 1 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, J.; Gao, T.; Wan, S.; Zhang, Y.; Yang, J.; Yu, Y.; Wang, W. Genome-Wide Identification, Classification and Expression Analysis of the HSP Gene Superfamily in Tea Plant (Camellia sinensis). Int. J. Mol. Sci. 2018, 19, 2633. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19092633
Chen J, Gao T, Wan S, Zhang Y, Yang J, Yu Y, Wang W. Genome-Wide Identification, Classification and Expression Analysis of the HSP Gene Superfamily in Tea Plant (Camellia sinensis). International Journal of Molecular Sciences. 2018; 19(9):2633. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19092633
Chicago/Turabian StyleChen, Jiangfei, Tong Gao, Siqing Wan, Yongheng Zhang, Jiankun Yang, Youben Yu, and Weidong Wang. 2018. "Genome-Wide Identification, Classification and Expression Analysis of the HSP Gene Superfamily in Tea Plant (Camellia sinensis)" International Journal of Molecular Sciences 19, no. 9: 2633. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19092633