ALLENE OXIDE SYNTHASE and HYDROPEROXIDE LYASE, Two Non-Canonical Cytochrome P450s in Arabidopsis thaliana and Their Different Roles in Plant Defense
Abstract
:1. Introduction
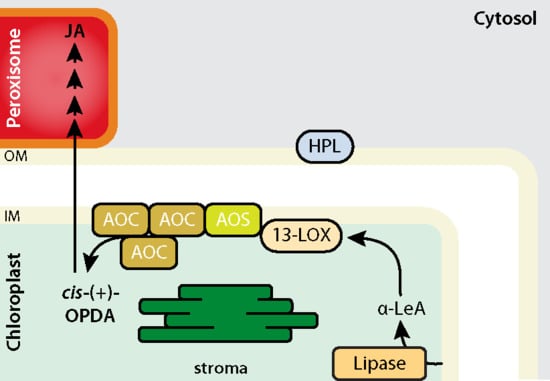
2. AOS and HPL Are Examples of Non-Canonical Cytochrome P450 Enzymes with Unique Activities in Chloroplasts
3. Structural Modelling of the LOX2–AOS–AOC2 Plastid Envelope Complex
4. Expression of LOX2, AOS, and AOC over Plant Development and Role of AOC Homo- and Hetero-Trimerization for Activity Regulation
5. Conclusions and Perspectives
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| 13-HPOT | (9Z11E15Z13S)-13-hydroperoxy-9,11,15-octadecatrienoic acid |
| α-LeA | α-linolenic acid |
| AOC | Allene oxide cyclase |
| AOS | Allene oxide synthase |
| CYP | Cytochrome P450 enzymes |
| EOT | 12,13(S)-epoxy-9(Z),11,15(Z)-octadecatrienoic acid |
| HPL | Hydroperoxide lyase |
| JA | Jasmonic acid |
| LOX | Lipoxygenase |
| ODA | 12-oxo-cis-9-dodecenoic acid |
| OPDA | cis-(+)-12-oxophytodienoic acid |
References
- Peters-Golden, M.; Brock, T.G. Intracellular compartmentalization of leukotriene biosynthesis. Am. J. Respir. Crit. Care Med. 2000, 161, S36–S40. [Google Scholar] [CrossRef] [PubMed]
- Soberman, R.J.; Christmas, P. The organization and consequences of eicosanoid signaling. J. Clin. Investig. 2003, 111, 1107–1113. [Google Scholar] [CrossRef] [PubMed]
- Sharma, J.N.; Mohammed, L.A. The role of leukotrienes in the pathophysiology of inflammatory disorders: Is there a case for revisiting leukotrienes as therapeutic targets? Inflammopharmacology 2006, 14, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Corey, E.J.; D’Alarcao, M.; Matsuda, S.P.T.; Lansbury, P.T., Jr.; Yamada, Y. Intermediacy of 8-(R)-HPETE in the conversion of arachidonic acid to pre-clavulone a by Clavularia viridis. Implications for the biosynthesis of marine prostanoids. J. Am. Chem. Soc. 1987, 109, 289–290. [Google Scholar] [CrossRef]
- Corey, E.J.; Matsuda, S.P.T.; Nagata, R.; Cleaver, M.B. Biosynthesis of 8-R-HPETE and preclavulone-A from arachidonate in several species of caribbean coral. A widespread route to marine prostanoids. Tetrahedron Lett. 1988, 29, 2555–2558. [Google Scholar] [CrossRef]
- Tijet, N.; Brash, A.R. Allene oxide synthases and allene oxides. Prostaglandins Other Lipid Medi. 2002, 68–69, 423–431. [Google Scholar] [CrossRef]
- Wasternack, C.; Strnad, M. Jasmonates: News on occurrence, biosynthesis, metabolism and action of an ancient group of signaling compounds. Int. J. Mol. Sci. 2018, 19, 2539. [Google Scholar] [CrossRef]
- Reinbothe, C.; Springer, A.; Samol, I.; Reinbothe, S. Plant oxylipins: Role of jasmonic acid during programmed cell death, defence and leaf senescence. FEBS J. 2009, 276, 4666–4681. [Google Scholar] [CrossRef]
- Yan, Y.; Borrego, E.; Kolomiets, M.V. Jasmonate biosynthesis, perception and function in plant development and stress responses. In Lipid Metabolism; Valenzuela Baez, R.E., Ed.; InTechOpen Limited: London, UK, 2013; pp. 393–442. [Google Scholar]
- Wasternack, C.; Feussner, I. The oxylipin pathways: Biochemistry and function. Annu. Rev. Plant Biol. 2018, 69, 363–386. [Google Scholar] [CrossRef]
- Schommer, C.; Palatnik, J.F.; Aggarwal, P.; Chetelat, A.; Cubas, P.; Farmer, E.E.; Nath, U.; Weigel, D. Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol 2008, 6, e230. [Google Scholar] [CrossRef]
- Hoffmann, M.; Hentrich, M.; Pollmann, S. Auxin-oxylipin crosstalk: Relationship of antagonists. J. Integr. Plant Biol. 2011, 53, 429–445. [Google Scholar] [CrossRef] [PubMed]
- Gfeller, A.; Liechti, R.; Farmer, E.E. Arabidopsis jasmonate signaling pathway. Sci. Signal 2010, 3, cm4. [Google Scholar] [CrossRef] [PubMed]
- Farmer, E.E.; Ryan, C.A. Interplant communication: Airborne methyl jasmonate induces synthesis of proteinase inhibitors in plant leaves. Proc. Natl. Acad. Sci. USA 1990, 87, 7713–7716. [Google Scholar] [CrossRef] [PubMed]
- Mattiacci, L.; Dicke, M.; Posthumus, M.A. ß-Glucosidase: An elicitor of herbivore-induced plant odor that attracts host-searching parasitic wasps. Proc. Natl. Acad. Sci. USA 1995, 92, 2036–2040. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, I.T. Functional interactions in the use of direct and indirect defences in native Nicotiana plants. Novartis Found Symp. 1999, 223, 74–87. [Google Scholar] [PubMed]
- Kessler, A.; Baldwin, I.T. Defensive function of herbivore-induced plant volatile emissions in nature. Science 2001, 291, 2141–2144. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wang, L.; Baldwin, I.T. Methyl jasmonate-elicited herbivore resistance: Does MeJA function as a signal without being hydrolyzed to JA? Planta 2008, 227, 1161–1168. [Google Scholar] [CrossRef]
- Arimura, G.; Matsui, K.; Takabayashi, J. Chemical and molecular ecology of herbivore-induced plant volatiles: Proximate factors and their ultimate functions. Plant Cell Physiol. 2009, 50, 911–923. [Google Scholar] [CrossRef]
- Arimura, G.; Shiojiri, K.; Karban, R. Acquired immunity to herbivory and allelopathy caused by airborne plant emissions. Phytochemistry 2010, 71, 1642–1649. [Google Scholar] [CrossRef]
- Wu, J.; Baldwin, I.T. New insights into plant responses to the attack from insect herbivores. Annu. Rev. Genet. 2010, 44, 1–24. [Google Scholar] [CrossRef]
- ul Hassan, M.N.; Zainal, Z.; Ismail, I. Green leaf volatiles: Biosynthesis, biological functions and their applications in biotechnology. Plant Biotechnol. J. 2015, 13, 727–739. [Google Scholar] [CrossRef] [PubMed]
- Matsui, K.; Koeduka, T. Green leaf volatiles in plant signaling and response. In Lipids in Plant and Algae Development, Subcellular Biochemistry; Nakamura, Y., Li-Beisson, Y., Eds.; Springer: Berlin/Heidelberg, Germany, 2016; Volume 86, pp. 427–443. [Google Scholar]
- Ameye, M.; Allmann, S.; Verwaeren, J.; Smagghe, G.; Haesaert, G.; Schuurink, R.C.; Audenaert, K. Green leaf volatile production by plants: A meta-analysis. New Phytol. 2018, 220, 666–683. [Google Scholar] [CrossRef] [PubMed]
- Schaller, A.; Stintzi, A. Enzymes in jasmonate biosynthesis—Structure, function, regulation. Phytochemistry 2009, 70, 1532–1538. [Google Scholar] [CrossRef] [PubMed]
- Gfeller, A.; Dubugnon, L.; Liechti, R.; Farmer, E.E. Jasmonate biochemical pathway. Sci. Signal 2010, 3, cm3. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, G. Biosynthesis and analysis of plant oxylipins. Free Radic. Res. 2015, 49, 565–582. [Google Scholar] [CrossRef] [PubMed]
- Vick, B.A.; Zimmerman, D.C. Biosynthesis of jasmonic acid by several plant species. Plant Physiol. 1984, 75, 458–461. [Google Scholar] [CrossRef] [PubMed]
- Vick, B.A.; Zimmerman, D.C. Pathways of fatty acid hydroperoxide metabolism in spinach leaf chloroplasts. Plant Physiol. 1987, 85, 1073–1078. [Google Scholar] [CrossRef]
- Feussner, I.; Wasternack, C. The lipoxygenase pathway. Annu. Rev. Plant Biol. 2002, 53, 275–297. [Google Scholar] [CrossRef]
- Laudert, D.; Pfannschmidt, U.; Lottspeich, F.; Holländer-Czytko, H.; Weiler, E.W. Cloning, molecular and functional characterization of Arabidopsis thaliana allene oxide synthase (CYP 74), the first enzyme of the octadecanoid pathway to jasmonates. Plant Mol. Biol. 1996, 31, 323–335. [Google Scholar] [CrossRef]
- Hofmann, E.; Zerbe, P.; Schaller, F. The crystal structure of Arabidopsis thaliana allene oxide cyclase: Insights into the oxylipin cyclization reaction. Plant Cell 2006, 18, 3201–3217. [Google Scholar] [CrossRef]
- Schaller, F.; Zerbe, P.; Reinbothe, S.; Reinbothe, C.; Hofmann, E.; Pollmann, S. The allene oxide cyclase family of Arabidopsis thaliana: Localization and cyclization. FEBS J. 2008, 275, 2428–2441. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, E.; Pollmann, S. Molecular mechanism of enzymatic allene oxide cyclization in plants. Plant Physiol. Biochem. 2008, 46, 302–308. [Google Scholar] [CrossRef] [PubMed]
- Joo, Y.C.; Oh, D.K. Lipoxygenases: Potential starting biocatalysts for the synthesis of signaling compounds. Biotechnol. Adv. 2012, 30, 1524–1532. [Google Scholar] [CrossRef] [PubMed]
- Croft, K.; Jüttner, F.; Slusarenko, A.J. Volatile Products of the Lipoxygenase Pathway Evolved from Phaseolus vulgaris (L.) Leaves Inoculated with Pseudomonas syringae pv phaseolicola. Plant Physiol. 1993, 101, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Bate, N.J.; Rothstein, S.J. C6-volatiles derived from the lipoxygenase pathway induce a subset of defense-related genes. Plant J. 1998, 16, 561–569. [Google Scholar] [CrossRef] [PubMed]
- Blée, E. Phytooxylipins and plant defense reactions. Prog. Lipid Res. 1998, 37, 33–72. [Google Scholar] [CrossRef]
- Böttcher, C.; Pollmann, S. Plant oxylipins: Plant responses to 12-oxo-phytodienoic acid are governed by its specific structural and functional properties. FEBS J. 2009, 276, 4693–4704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 2000, 408, 796–815. [Google Scholar] [CrossRef]
- Song, W.C.; Brash, A.R. Purification of an allene oxide synthase and identification of the enzyme as a cytochrome P-450. Science 1991, 253, 781–784. [Google Scholar] [CrossRef]
- Froehlich, J.E.; Itoh, A.; Howe, G.A. Tomato allene oxide synthase and fatty acid hydroperoxide lyase, two cytochrome P450s involved in oxylipin metabolism, are targeted to different membranes of chloroplast envelope. Plant Physiol. 2001, 125, 306–317. [Google Scholar] [CrossRef]
- La Camera, S.; Gouzerh, G.; Dhondt, S.; Hoffmann, L.; Fritig, B.; Legrand, M.; Heitz, T. Metabolic reprogramming in plant innate immunity: The contributions of phenylpropanoid and oxylipin pathways. Immunol. Rev. 2004, 198, 267–284. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.S.; Nioche, P.; Hamberg, M.; Raman, C.S. Structural insights into the evolutionary paths of oxylipin biosynthetic enzymes. Nature 2008, 455, 363–368. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Chang, Z.; Pan, Z.; Fu, Z.Q.; Wang, X. Modes of heme binding and substrate access for cytochrome P450 CYP74A revealed by crystal structures of allene oxide synthase. Proc. Natl. Acad. Sci. USA 2008, 105, 13883–13888. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, P.A.; Cosme, J.; Sridhar, V.; Johnson, E.F.; McRee, D.E. Microsomal cytochrome P450 2C5: Comparison to microbial P450s and unique features. J. Inorg. Biochem. 2000, 81, 183–190. [Google Scholar] [CrossRef]
- Tyagi, C.; Singh, A.; Singh, I.K. Mechanistic insights into mode of action of rice allene oxide synthase on hydroxyperoxides: An intermediate step in herbivory-induced jasmonate pathway. Comput. Biol. Chem. 2016, 64, 227–236. [Google Scholar] [CrossRef] [PubMed]
- Williams, P.A.; Cosme, J.; Sridhar, V.; Johnson, E.F.; McRee, D.E. Mammalian microsomal cytochrome P450 monooxygenase: Structural adaptations for membrane binding and functional diversity. Mol. Cell 2000, 5, 121–131. [Google Scholar] [CrossRef]
- Nelson, D.R. Plant cytochrome P450s from moss to poplar. Phytochem. Rev. 2006, 5, 193–204. [Google Scholar] [CrossRef] [Green Version]
- Grechkin, A.N.; Hamberg, M. The “heterolytic hydroperoxide lyase” is an isomerase producing a short-lived fatty acid hemiacetal. Biochim. Biophys. Acta 2004, 1636, 47–58. [Google Scholar] [CrossRef]
- Hooper, C.M.; Castleden, I.R.; Tanz, S.K.; Aryamanesh, N.; Millar, A.H. SUBA4: The interactive data analysis centre for Arabidopsis subcellular protein locations. Nucleic Acids Res. 2017, 45, D1064–D1074. [Google Scholar] [CrossRef]
- Froehlich, J.E.; Wilkerson, C.G.; Ray, W.K.; McAndrew, R.S.; Osteryoung, K.W.; Gage, D.A.; Phinney, B.S. Proteomic study of the Arabidopsis thaliana chloroplastic envelope membrane utilizing alternatives to traditional two-dimensional electrophoresis. J. Proteome Res. 2003, 2, 413–425. [Google Scholar] [CrossRef]
- Pollmann, S.; Springer, A.; Rustgi, S.; Wettstein, D.V.; Kang, C.; Reinbothe, C.; Reinbothe, S. Substrate channeling in oxylipin biosynthesis through a protein complex in the plastid envelope of Arabidopsis thaliana. J. Exp. Bot. 2019, 70, 1483–1495. [Google Scholar] [CrossRef] [PubMed]
- Emanuelsson, O.; Nielsen, H.; von Heijne, G. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 1999, 8, 978–984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emanuelsson, O.; Nielsen, H.; Brunak, S.; von Heijne, G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000, 300, 1005–1016. [Google Scholar] [CrossRef] [PubMed]
- Farmaki, T.; Sanmartin, M.; Jimenez, P.; Paneque, M.; Sanz, C.; Vancanneyt, G.; Leon, J.; Sanchez-Serrano, J.J. Differential distribution of the lipoxygenase pathway enzymes within potato chloroplasts. J. Exp. Bot. 2007, 58, 555–568. [Google Scholar] [CrossRef] [PubMed]
- Corey, E.J.; Washburn, W.N.; Chen, J.C. Prostaglandin A2 synthetase complex from Plexaura homomalla. J. Am. Chem. Soc. 1973, 95, 2054–2055. [Google Scholar] [CrossRef] [PubMed]
- Koljak, R.; Boutaud, O.; Shieh, B.H.; Samel, N.; Brash, A.R. Identification of a naturally occurring peroxidase-lipoxygenase fusion protein. Science 1997, 277, 1994–1996. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, N.C.; Niebuhr, M.; Tsuruta, H.; Bordelon, T.; Ridderbusch, O.; Dassey, A.; Brash, A.R.; Bartlett, S.G.; Newcomer, M.E. A covalent linker allows for membrane targeting of an oxylipin biosynthetic complex. Biochemistry 2008, 47, 10665–10676. [Google Scholar] [CrossRef]
- Oldham, M.L.; Brash, A.R.; Newcomer, M.E. Insights from the X-ray crystal structure of coral 8R-lipoxygenase: Calcium activation via a C2-like domain and a structural basis of product chirality. J. Biol. Chem. 2005, 280, 39545–39552. [Google Scholar] [CrossRef]
- Oldham, M.L.; Brash, A.R.; Newcomer, M.E. The structure of coral allene oxide synthase reveals a catalase adapted for metabolism of a fatty acid hydroperoxide. Proc. Natl. Acad. Sci. USA 2005, 102, 297–302. [Google Scholar] [CrossRef]
- Youn, B.; Sellhorn, G.E.; Mirchel, R.J.; Gaffney, B.J.; Grimes, H.D.; Kang, C. Crystal structures of vegetative soybean lipoxygenase VLX-B and VLX-D, and comparisons with seed isoforms LOX-1 and LOX-3. Proteins Struct. Funct. Bioinform. 2006, 65, 1008–1020. [Google Scholar] [CrossRef] [Green Version]
- Rizo, J.; Südhof, T.C. C2-domains, structure and function of a universal Ca2+-binding domain. J. Biol. Chem. 1998, 273, 15879–15882. [Google Scholar] [CrossRef] [PubMed]
- Cho, W.; Stahelin, R.V. Membrane binding and subcellular targeting of C2 domains. Biochim. Biophys. Acta 2006, 1761, 838–849. [Google Scholar] [CrossRef] [PubMed]
- Skrzypczak-Jankun, E.; Amzel, L.M.; Kroa, B.A.; Funk, M.O., Jr. Structure of soybean lipoxygenase L3 and a comparison with its L1 isoenzyme. Proteins 1997, 29, 15–31. [Google Scholar] [CrossRef]
- Schwede, T.; Kopp, J.; Guex, N.; Peitsch, M.C. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003, 31, 3381–3385. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, K.; Stoffel, W. Tmbase—A database of membrane spanning proteins segments. Biol. Chem. Hoppe-Seyler 1993, 374, 166. [Google Scholar]
- Brock, T.G.; Healy, A.M. Nuclear import of arachidonate 5-lipoxygenase. Arch. Immunol. Ther. Exp. 2000, 48, 481–486. [Google Scholar]
- Chen, X.S.; Funk, C.D. The N-terminal “beta-barrel” domain of 5-lipoxygenase is essential for nuclear membrane translocation. J. Biol. Chem. 2001, 276, 811–818. [Google Scholar] [CrossRef] [PubMed]
- Schewe, T.; Rapoport, S.M.; Kuhn, H. Enzymology and physiology of reticulocyte lipoxygenase: Comparison with other lipoxygenases. Adv. Enzymol. Relat. Areas Mol. Biol. 1986, 58, 191–272. [Google Scholar]
- van Leyen, K.; Duvoisin, R.M.; Engelhardt, H.; Wiedmann, M. A function for lipoxygenase in programmed organelle degradation. Nature 1998, 395, 392–395. [Google Scholar] [CrossRef]
- Cook, A.C.; Ho, C.; Kershner, J.L.; Malinowski, S.A.; Moldveen, H.; Stagliano, B.A.; Slater, S.J. Competitive binding of protein kinase Calpha to membranes and Rho GTPases. Biochemistry 2006, 45, 14452–14465. [Google Scholar] [CrossRef]
- Otto, M.; Naumann, C.; Brandt, W.; Wasternack, C.; Hause, B. Activity Regulation by heteromerization of Arabidopsis allene oxide cyclase family members. Plants 2016, 5, 3. [Google Scholar] [CrossRef] [PubMed]
- Hruz, T.; Laule, O.; Szabo, G.; Wessendorp, F.; Bleuler, S.; Oertle, L.; Widmayer, P.; Gruissem, W.; Zimmermann, P. Genevestigator v3: A reference expression database for the meta-analysis of transcriptomes. Adv. Bioinform. 2008, 2008, 420747. [Google Scholar] [CrossRef] [PubMed]
- Bell, C.D.; Soltis, D.E.; Soltis, P.S. The age of the angiosperms: A molecular timescale without a clock. Evolution 2005, 59, 1245–1258. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.; Niisuke, K.; Boeglin, W.E.; Voehler, M.; Stec, D.F.; Porter, N.A.; Brash, A.R. Enzymatic synthesis of a bicyclobutane fatty acid by a hemoprotein lipoxygenase fusion protein from the cyanobacterium Anabaena PCC 7120. Proc. Natl. Acad. Sci. USA 2007, 104, 18941–18945. [Google Scholar] [CrossRef] [PubMed]
- Lang, I.; Gobel, C.; Porzel, A.; Heilmann, I.; Feussner, I. A lipoxygenase with linoleate diol synthase activity from Nostoc sp. PCC 7120. Biochem. J. 2008, 410, 347–357. [Google Scholar] [PubMed]
- Teder, T.; Lõhelaid, H.; Boeglin, W.E.; Calcutt, W.M.; Brash, A.R.; Samel, N. A Catalase-related hemoprotein in coral is specialized for synthesis of short-chain aldehydes: Discovery of P450-type hydroperoxide lyase activity in a catalase. J. Biol. Chem. 2015, 290, 19823–19832. [Google Scholar] [CrossRef] [PubMed]
- Teder, T.; Lõhelaid, H.; Samel, N. Structural and functional insights into the reaction specificity of catalase-related hydroperoxide lyase: A shift from lyase activity to allene oxide synthase by site-directed mutagenesis. PLoS ONE 2017, 12, e0185291. [Google Scholar] [CrossRef] [PubMed]
- Niisuke, K.; Boeglin, W.E.; Murray, J.J.; Schneider, C.; Brash, A.R. Biosynthesis of a linoleic acid allylic epoxide: Mechanistic comparison with its chemical synthesis and leukotriene A biosynthesis. J. Lipid Res. 2009, 50, 1448–1455. [Google Scholar] [CrossRef] [PubMed]
- Hecker, M.; Ullrich, V. On the mechanism of prostacyclin and thromboxane A2 biosynthesis. J. Biol. Chem. 1989, 264, 141–150. [Google Scholar] [PubMed]
- Gerwick, W.H. Epoxy allylic carbocations as conceptual intermediates in the biogenesis of diverse marine oxylipins. Lipids 1996, 31, 1215–1231. [Google Scholar] [CrossRef]
- Lõhelaid, H.; Teder, T.; Tõldsepp, K.; Ekins, M.; Samel, N. Up-regulated expression of AOS-LOXa and increased eicosanoid synthesis in response to coral wounding. PLoS ONE 2014, 9, e89215. [Google Scholar] [CrossRef] [PubMed]






| Amino Acid Interactions Observed in the Ternary Complex | Amino Acid Interactions Observed in an Arbitrary AOS–AOC2 Complex |
|---|---|
| LOX2-Ser92–AOS-Ser272 | AOS-Pro64–AOC2-Ser45 |
| LOX2-Gly94–AOS-Ser272 LOX2-Asp96–AOC2-Asn42 | AOS-Ile65–AOC2-Phe44 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rustgi, S.; Springer, A.; Kang, C.; von Wettstein, D.; Reinbothe, C.; Reinbothe, S.; Pollmann, S. ALLENE OXIDE SYNTHASE and HYDROPEROXIDE LYASE, Two Non-Canonical Cytochrome P450s in Arabidopsis thaliana and Their Different Roles in Plant Defense. Int. J. Mol. Sci. 2019, 20, 3064. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20123064
Rustgi S, Springer A, Kang C, von Wettstein D, Reinbothe C, Reinbothe S, Pollmann S. ALLENE OXIDE SYNTHASE and HYDROPEROXIDE LYASE, Two Non-Canonical Cytochrome P450s in Arabidopsis thaliana and Their Different Roles in Plant Defense. International Journal of Molecular Sciences. 2019; 20(12):3064. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20123064
Chicago/Turabian StyleRustgi, Sachin, Armin Springer, ChulHee Kang, Diter von Wettstein, Christiane Reinbothe, Steffen Reinbothe, and Stephan Pollmann. 2019. "ALLENE OXIDE SYNTHASE and HYDROPEROXIDE LYASE, Two Non-Canonical Cytochrome P450s in Arabidopsis thaliana and Their Different Roles in Plant Defense" International Journal of Molecular Sciences 20, no. 12: 3064. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20123064