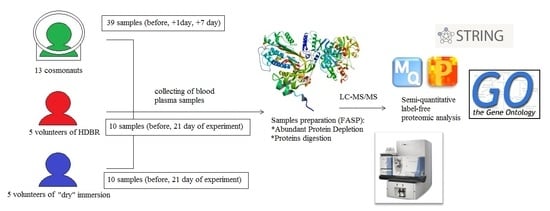
The Effects of Spaceflight Factors on the Human Plasma Proteome, Including Both Real Space Missions and Ground-Based Experiments
Abstract
:1. Introduction
2. Results and Discussion
2.1. Proteomic Analysis of Blood Plasma in Ground-Based Experiments and Spaceflight
2.1.1. Head-Down Bed Rest (HDBR)
2.1.2. Dry Immersion
2.2. Spaceflight
2.3. Comparison of Ground-Based Experiments and Spaceflight
3. Materials and Methods
3.1. Sample Collection
3.2. LC-MS/MS Proteomic Analysis
3.3. Data Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kaur, I.; Simons, E.R.; Castro, V.A.; Mark Ott, C.; Pierson, D.L. Changes in neutrophil functions in astronauts. Brain. Behav. Immun. 2004, 18, 443–450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, D.; Kuipers, A.; Mukai, C.; Thirsk, R. Acclimation during space flight: Effects on human physiology. CMAJ 2009, 180, 1317–1323. [Google Scholar] [CrossRef] [PubMed]
- Grimm, D.; Pietsch, J.; Richter, M.; Peter, W.; Strauch, S.M.; Lebert, M.; Magnusson, N.E.; Wise5, P.; Bauer, J. The impact of microgravity-based proteomics research. Expert Rev. Proteom. 2014, 11, 465–475. [Google Scholar] [CrossRef] [PubMed]
- Murata, Y.; Yasuda, T.; Watanabe-Asaka, T.; Oda, S.; Mantoku, A.; Takeyama, K.; Chatani, M.; Kudo, A.; Uchida, S.; Suzuki, H.; et al. Histological and transcriptomic analysis of adult Japanese Medaka sampled onboard the international space station. PLoS ONE 2015, 10, e0138799. [Google Scholar] [CrossRef]
- Stein, T.P. Weight, muscle and bone loss during space flight: Another perspective. Eur. J. Appl. Physiol. 2013, 113, 2171–2181. [Google Scholar] [CrossRef] [PubMed]
- Pastushkova, L.H.; Rusanov, V.B.; Goncharova, A.G.; Brzhozovskiy, A.G.; Kononikhin, A.S.; Chernikova, A.G.; Kashirina, D.N.; Nosovsky, A.M.; Baevsky, R.M.; Nikolaev, E.N.; et al. Urine proteome changes associated with autonomic regulation of heart rate in cosmonauts. BMC Syst. Biol. 2018, 13, 17. [Google Scholar] [CrossRef]
- Hatton, D.C.; Yue, Q.; Dierickx, J.; Roullet, C.; Otsuka, K.; Watanabe, M.; Coste, S.; Roullet, J.B.; Phanouvang, T.; Orwoll, E.; et al. Calcium metabolism and cardiovascular function after spaceflight. J. Appl. Physiol. 2002, 92, 3–12. [Google Scholar] [CrossRef]
- Watanabe, Y.; Ohshima, H.; Mizuno, K.; Sekiguchi, C.; Fukunaga, M.; Kohri, K.; Rittweger, J.; Felsenberg, D.; Matsumoto, T.; Nakamura, T. Intravenous pamidronate prevents femoral bone loss and renal stone formation during 90-day bed rest. J. Bone Miner. Res. 2004, 19, 1771–1778. [Google Scholar] [CrossRef]
- Linossier, M.-T.; Amirova, L.E.; Thomas, M.; Normand, M.; Bareille, M.; Gauquelin-koch, G.; Beck, A.; Bonneau, C.; Gharib, C.; Custaud, M.; et al. Effects of short-term dry immersion on bone remodeling markers, insulin and adipokines. PLoS ONE 2017, 12, e0182970. [Google Scholar] [CrossRef]
- Biolo, G.; Ciocchi, B.; Lebenstedt, M.; Barazzoni, R.; Zanetti, M.; Platen, P.; Heer, M.; Guarnieri, G. Short-term bed rest impairs amino acid-induced protein anabolism in humans. J. Physiol. 2004, 558, 381–388. [Google Scholar] [CrossRef] [Green Version]
- Ulbrich, C.; Wehland, M.; Pietsch, J.; Aleshcheva, G.; Wise, P.; Van Loon, J.; Magnusson, N.; Infanger, M.; Grosse, J.; Eilles, C.; et al. The impact of simulated and real microgravity on bone cells and mesenchymal stem cells. BioMed Res. Int. 2014, 2014. [Google Scholar] [CrossRef] [PubMed]
- Kaur, I.; Simons, E.R.; Castro, V.A.; Ott, C.M.; Pierson, D.L. Changes in monocyte functions of astronauts. Brain. Behav. Immun. 2005, 19, 547–554. [Google Scholar] [CrossRef] [PubMed]
- Ferrando, A.A.; Paddon-Jones, D.; Wolfe, R.R. Alterations in protein metabolism during space flight and inactivity. Nutrition 2002, 18, 837–841. [Google Scholar] [CrossRef]
- Grosse, J.; Wehland, M.; Pietsch, J.; Ma, X.; Ulbrich, C.; Schulz, H.; Saar, K.; Hubner, N.; Hauslage, J.; Hemmersbach, R.; et al. Short-term weightlessness produced by parabolic flight maneuvers altered gene expression patterns in human endothelial cells. FASEB J. 2012, 26, 639–655. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Han, D.K. Overcoming the dynamic range problem in mass spectrometry-based shotgun proteomics. Expert Rev. Proteom. 2006, 3, 611–619. [Google Scholar] [CrossRef] [PubMed]
- Binder, H.; Wirth, H.; Arakelyan, A.; Lembcke, K.; Tiys, E.S.; Ivanisenko, V.A.; Kolchanov, N.A.; Kononikhin, A.; Popov, I.; Nikolaev, E.N.; et al. Time-course human urine proteomics in space-flight simulation experiments. BMC Genom. 2014, 15, S2. [Google Scholar] [CrossRef] [PubMed]
- Kopylov, A.T.; Ilgisonis, E.V.; Moysa, A.A.; Tikhonova, O.V.; Zavialova, M.G.; Novikova, S.E.; Lisitsa, A.V.; Ponomarenko, E.A.; Moshkovskii, S.A.; Markin, A.A.; et al. Targeted Quantitative Screening of Chromosome 18 Encoded Proteome in Plasma Samples of Astronaut Candidates. J. Proteome Res. 2016, 15, 4039–4046. [Google Scholar] [CrossRef]
- Khristenko, N.A.; Larina, I.M.; Domon, B. Longitudinal Urinary Protein Variability in Participants of the Space Flight Simulation Program. J. Proteome Res. 2016, 15, 114–124. [Google Scholar] [CrossRef]
- Kononikhin, A.S.; Starodubtseva, N.L.; Pastushkova, L.K.; Kashirina, D.N.; Fedorchenko, K.Y.; Brhozovsky, A.G.; Popov, I.A.; Larina, I.M.; Nikolaev, E.N. Spaceflight induced changes in the human proteome. Expert Rev. Proteom. 2017, 14, 15–29. [Google Scholar] [CrossRef]
- Moriggi, M.; Vasso, M.; Fania, C.; Capitanio, D.; Bonifacio, G.; Salanova, M.; Blottner, D.; Rittweger, J.; Felsenberg, D.; Cerretelli, P.; et al. Long term bed rest with and without vibration exercise countermeasures: Effects on human muscle protein dysregulation. Proteomics 2010, 10, 3756–3774. [Google Scholar] [CrossRef] [Green Version]
- Meloni, M.A.; Galleri, G.; Pani, G.; Saba, A.; Pippia, P.; Cogoli-Greuter, M. Space flight affects motility and cytoskeletal structures in human monocyte cell line J-111. Cytoskeleton 2011, 68, 125–137. [Google Scholar] [CrossRef] [PubMed]
- Nelson, E.; Mulugeta, L.; Myers, J. Microgravity-Induced Fluid Shift and Ophthalmic Changes. Life 2014, 4, 621–665. [Google Scholar] [CrossRef] [PubMed]
- Delp, M.D.; Charvat, J.M.; Limoli, C.L.; Globus, R.K.; Ghosh, P. Apollo Lunar Astronauts Show Higher Cardiovascular Disease Mortality: Possible Deep Space Radiation Effects on the Vascular Endothelium. Sci. Rep. 2016, 6, 29901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, S.M. Red blood cell and iron metabolism during space flight. Nutrition 2002, 18, 864–866. [Google Scholar] [CrossRef]
- Gresele, P.; Kleiman, N.S.; Lopez, J.A.; Page, C.P. Platelets in Thrombotic and Non-thrombotic Disorders: Pathophysiology, Pharmacology and Therapeutics: An Update; Springer: New York, NY, USA, 2017; ISBN 978-3-319-47460-1. [Google Scholar]
- Canobbio, I.; Oliviero, B.; Pula, G.; Vara, D.; Guidetti, G.F.; Manganaro, D.; Torti, M. Amyloid β-peptide-dependent activation of human platelets: Essential role for Ca 2+ and ADP in aggregation and thrombus formation. Biochem. J. 2014, 462, 513–523. [Google Scholar] [CrossRef] [PubMed]
- Crucian, B.; Sams, C. Immune system dysregulation during spaceflight: Clinical risk for exploration-class missions. J. Leukoc. Biol. 2009, 86, 1017–1018. [Google Scholar] [CrossRef] [PubMed]
- Cogoli, A.; Tschopp, A.; Fuchs-Bislin, P. Cell Sensitivity to Gravity. Science 1984, 225, 228–230. [Google Scholar] [CrossRef]
- Crucian, B.; Stowe, R.; Quiriarte, H.; Pierson, D.; Sams, C. Monocyte phenotype and cytokine production profiles are dysregulated by short-duration spaceflight. Aviat. Space Environ. Med. 2011, 82, 857–862. [Google Scholar] [CrossRef]
- Misumi, Y.; Ando, Y.; Gonçalves, N.P.; Saraiva, M.J. Fibroblasts endocytose and degrade transthyretin aggregates in transthyretin-related amyloidosis. Lab. Investig. 2013, 93, 911–920. [Google Scholar] [CrossRef] [Green Version]
- Whinnery, J.E. Comparative distribution of petechial haemorrhages as a function of aircraft cockpit geometry. J. Biomed. Eng. 1987, 9, 201–205. [Google Scholar] [CrossRef]
- Ganse, B.; Limper, U.; Bühlmeier, J.; Rittweger, J. Petechiae: Reproducible pattern of distribution and increased appearance after bed rest. Aviat. Space Environ. Med. 2013, 84, 864–866. [Google Scholar] [CrossRef] [PubMed]
- Pastushkova, L.K.; Kashirina, D.N.; Kononikhin, A.S.; Brzhozovsky, A.G.; Ivanisenko, V.A.; Tiys, E.S.; Novosyolova, A.M.; Custaud, M.-A.; Nikolaev, E.N.; Larina, I.M. The Effect of Long-term Space Flights on Human Urine Proteins Functionally Related to Endothelium. Hum. Physiol. 2018, 44, 60–67. [Google Scholar] [CrossRef]
- Cvirn, G.; Waha, J.E.; Ledinski, G.; Schlagenhauf, A.; Leschnik, B.; Koestenberger, M.; Tafeit, E.; Hinghofer-Szalkay, H.; Goswami, N. Bed rest does not induce hypercoagulability. Eur. J. Clin. Invest. 2015, 45, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016, 11, 2301–2319. [Google Scholar] [CrossRef] [PubMed]
- von Mering, C.; Huynen, M.; Jaeggi, D.; Schmidt, S.; Bork, P.; Snel, B. STRING: A database of predicted functional associations between proteins. Nucl. Acids Res. 2003, 31, 258–261. [Google Scholar] [CrossRef]
- Vizca, J.A.; Csordas, A.; Griss, J.; Lavidas, I.; Mayer, G.; Perez-riverol, Y.; Reisinger, F.; Ternent, T.; Xu, Q.; Wang, R.; et al. 2016 update of the PRIDE database and its related tools. Nucl. Acids Res. 2016, 44, 447–456. [Google Scholar] [CrossRef]




| Protein IDs | Gene Names | Protein Names | Unique Peptides | p-Value | t-Test Difference | HDBR 21 Day | Background | |
|---|---|---|---|---|---|---|---|---|
| Q9Y490 | TLN1 | Talin-1 | 46 | 8.4 × 10−6 | −4.6 | ˅ | 16.6 ± 0.6 | 21.2 ± 1.8 |
| P01861 | IGHG4 | Ig gamma-4 chain C region | 3 | 5.2 × 10−7 | −4.2 | ˅ | 18.5 ± 0.6 | 22.6 ± 1.8 |
| P02655 | APOC2 | Apolipoprotein C-II;Proapolipoprotein C-II | 5 | 9.9 × 10−6 | −3.6 | ˅ | 19.4 ± 1.2 | 23.1 ± 1.2 |
| P12814 | ACTN1 | Alpha-actinin-1 | 24 | 1.2 × 10−6 | −3.3 | ˅ | 16.7 ± 0.5 | 20 ± 2.3 |
| P01834 | IGKC | Ig kappa chain C region | 5 | 6.7 × 10−4 | −2.5 | ˅ | 19.3 ± 1.6 | 21.8 ± 3 |
| P63261; P60709 | ACTG1; ACTB | Actin. cytoplasmic 2; Actin. cytoplasmic 1 | 6 | 2.9 × 10−4 | −2.1 | ˅ | 18.4 ± 0.6 | 20.6 ± 2.5 |
| O00187 | MASP2 | Mannan-binding lectin serine protease 2 | 6 | 1.5 × 10−5 | −1.9 | ˅ | 18.6 ± 1.1 | 20.6 ± 1.1 |
| O00391 | QSOX1 | Sulfhydryl oxidase 1 | 13 | 1.3 × 10−4 | −1.4 | ˅ | 18.8 ± 0.6 | 20.2 ± 1.4 |
| P55058 | PLTP | Phospholipid transfer protein | 10 | 1.6 × 10−6 | −1.4 | ˅ | 18.1 ± 0.4 | 19.5 ± 0.8 |
| Q99878; Q96KK5; Q9BTM1; Q93077; Q8IUE6; Q7L7L0; P20671; P0C0S8; P04908; P16104 | HIST1H2AJ; HIST1H2AH; H2AFJ; HIST1H2AC; HIST2H2AB; HIST3H2A; HIST1H2AD; HIST1H2AG; HIST1H2AB; H2AFX | Histone H2A | 1 | 1.2 × 10−5 | −0.9 | ˅ | 17.2 ± 0.2 | 18.2 ± 0.9 |
| P05154 | SERPINA5 | Plasma serine protease inhibitor | 9 | 3.1 × 10−5 | 0.7 | ˄ | 19.5 ± 0.2 | 18.8 ± 0.5 |
| P07358 | C8B | Complement component C8 beta chain | 12 | 9.7 × 10−5 | 0.8 | ˄ | 20.5 ± 0.5 | 19.7 ± 1 |
| P03952 | KLKB1 | Plasma kallikrein | 9 | 1.3 × 10−4 | 0.9 | ˄ | 21.3 ± 0.6 | 20.4 ± 1 |
| P01024 | C3 | Complement C3 | 93 | 5.1 × 10−5 | 1.0 | ˄ | 27.2 ± 0.5 | 26.2 ± 1.2 |
| P22792 | CPN2 | Carboxypeptidase N subunit 2 | 6 | 6.8 × 10−4 | 1.1 | ˄ | 22 ± 0.6 | 21 ± 0.7 |
| P04114 | APOB | Apolipoprotein B-100 | 192 | 3.0 × 10−4 | 1.1 | ˄ | 26.1 ± 0.3 | 25.1 ± 1.7 |
| P06681 | C2 | Complement C2 | 12 | 2.6 × 10−4 | 1.2 | ˄ | 21.1 ± 0.4 | 19.9 ± 1.3 |
| P02766 | TTR | Transthyretin | 10 | 5.2 × 10−4 | 1.2 | ˄ | 23.1 ± 0.9 | 21.9 ± 1.7 |
| P25311 | AZGP1 | Zinc-alpha-2-glycoprotein | 13 | 5.6 × 10−6 | 1.4 | ˄ | 23 ± 0.6 | 21.6 ± 1.2 |
| P02774 | GC | Vitamin D-binding protein | 13 | 8.6 × 10−6 | 1.4 | ˄ | 23 ± 1 | 21.6 ± 1.3 |
| P04196 | HRG | Histidine-rich glycoprotein | 10 | 6.7 × 10−7 | 1.4 | ˄ | 23.5 ± 0.8 | 22.1 ± 1 |
| Q06033 | ITIH3 | Inter-alpha-trypsin inhibitor heavy chain H3 | 10 | 6.4 × 10−7 | 1.4 | ˄ | 20.9 ± 0.5 | 19.5 ± 1.2 |
| P01019 | AGT | Angiotensinogen | 10 | 2.7 × 10−5 | 1.4 | ˄ | 22 ± 1.1 | 20.6 ± 1.3 |
| P08519 | LPA | Apolipoprotein(a) | 7 | 4.8 × 10−4 | 1.5 | ˄ | 21.5 ± 1.3 | 20 ± 0.9 |
| P01023 | A2M | Alpha-2-macroglobulin | 36 | 2.0 × 10−6 | 1.5 | ˄ | 25.4 ± 0.6 | 23.9 ± 1.6 |
| P01042 | KNG1 | Kininogen-1 | 16 | 1.1 × 10−7 | 1.6 | ˄ | 23.1 ± 0.8 | 21.5 ± 1.1 |
| O75882 | ATRN | Attractin | 10 | 5.0 × 10−4 | 1.6 | ˄ | 21.3 ± 0.4 | 19.7 ± 1.5 |
| P00751 | CFB | Complement factor B | 18 | 6.8 × 10−8 | 1.7 | ˄ | 23.2 ± 0.5 | 21.5 ± 1.2 |
| P35858 | IGFALS | Insulin-like growth factor-binding protein complex acid labile subunit | 11 | 7.9 × 10−5 | 1.7 | ˄ | 20.9 ± 0.7 | 19.2 ± 1.6 |
| P05546 | SERPIND1 | Heparin cofactor 2 | 14 | 1.3 × 10−8 | 1.7 | ˄ | 22.6 ± 0.7 | 20.9 ± 0.9 |
| P02750 | LRG1 | Leucine-rich alpha-2-glycoprotein | 8 | 1.2 × 10−7 | 1.8 | ˄ | 22.4 ± 0.4 | 20.7 ± 1.2 |
| Q16610 | ECM1 | Extracellular matrix protein 1 | 9 | 1.7 × 10−4 | 1.8 | ˄ | 20.7 ± 0.5 | 18.9 ± 1.7 |
| Q14624 | ITIH4 | Inter-alpha-trypsin inhibitor heavy chain H4 | 37 | 3.2 × 10−9 | 1.9 | ˄ | 25.1 ± 0.4 | 23.2 ± 1.2 |
| P29622 | SERPINA4 | Kallistatin | 14 | 5.0 × 10−7 | 1.9 | ˄ | 21.5 ± 0.3 | 19.5 ± 1.5 |
| P08697 | SERPINF2 | Alpha-2-antiplasmin | 16 | 8.0 × 10−9 | 2.0 | ˄ | 22.6 ± 0.5 | 20.5 ± 1.4 |
| P01008 | SERPINC1 | Antithrombin-III | 22 | 9.4 × 10−8 | 2.1 | ˄ | 24.1 ± 0.8 | 22 ± 1.6 |
| P04217 | A1BG | Alpha-1B-glycoprotein | 12 | 1.0 × 10−7 | 2.1 | ˄ | 23.2 ± 0.9 | 21.1 ± 1.5 |
| P01009 | SERPINA1 | Alpha-1-antitrypsin | 23 | 4.9 × 10−8 | 2.2 | ˄ | 24.9 ± 1.2 | 22.7 ± 1.2 |
| P05155 | SERPING1 | Plasma protease C1 inhibitor | 14 | 7.4 × 10−7 | 2.3 | ˄ | 24 ± 1.3 | 21.7 ± 1.8 |
| P01011 | SERPINA3 | Alpha-1-antichymotrypsin | 19 | 1.6 × 10−9 | 2.6 | ˄ | 24.9 ± 1 | 22.2 ± 1.3 |
| P02790 | HPX | Hemopexin | 14 | 4.6 × 10−8 | 2.7 | ˄ | 25.9 ± 1.3 | 23.2 ± 1.8 |
| P19827 | ITIH1 | Inter-alpha-trypsin inhibitor heavy chain H1 | 24 | 4.1 × 10−9 | 2.8 | ˄ | 24.6 ± 0.6 | 21.8 ± 1.9 |
| P01031 | C5 | Complement C5 | 34 | 2.7 × 10−9 | 3.2 | ˄ | 23.6 ± 0.3 | 20.5 ± 1.9 |
| P19823 | ITIH2 | Inter-alpha-trypsin inhibitor heavy chain H2 | 32 | 7.3 × 10−10 | 3.2 | ˄ | 25.6 ± 0.6 | 22.4 ± 1.8 |
| Protein IDs | Gene Names | Protein Names | Unique Peptides | p-Value | t-Test Difference | Immersion 21 Day | Background | |
|---|---|---|---|---|---|---|---|---|
| P01024 | C3 | Complement C3 | 93 | 6.8 × 10−6 | 1.0 | ˄ | 27.2 ± 0.5 | 26.2 ± 1.2 |
| P02766 | TTR | Transthyretin | 10 | 1.3 × 10−5 | 1.1 | ˄ | 23 ± 0.4 | 21.9 ± 1.7 |
| P04114 | APOB | Apolipoprotein B-100; Apolipoprotein B-48 | 192 | 4.7 × 10−7 | 1.6 | ˄ | 25.4 ± 0.6 | 24.2 ± 1.5 |
| O75636 | FCN3 | Ficolin-3 | 10 | 9.1 × 10−6 | 1.7 | ˄ | 27.7 ± 1.1 | 26.4 ± 1.4 |
| P00450 | CP | Ceruloplasmin | 39 | 5.6 × 10−6 | 1.2 | ˄ | 23.7 ± 0.7 | 22.3 ± 1.7 |
| P00734 | F2 | Prothrombin | 10 | 3.1 × 10−5 | 1.7 | ˄ | 21.5 ± 0.6 | 20.1 ± 1.4 |
| P00736 | C1R | Complement C1r subcomponent | 14 | 7.5 × 10−6 | 2.4 | ˄ | 26.6 ± 0.3 | 25.1 ± 1.7 |
| P00738 | HP | Haptoglobin | 7 | 1.8 × 10−5 | 1.9 | ˄ | 22.1 ± 0.7 | 20.5 ± 2.1 |
| P01857 | IGHG1 | Ig gamma-1 chain C region | 4 | 2.2 × 10−6 | 2.6 | ˄ | 22.7 ± 0.4 | 21 ± 2.1 |
| P01871; P04220 | IGHM | Ig mu chain C region | 11 | 4.3 × 10−6 | 2.0 | ˄ | 24.7 ± 0.9 | 22.9 ± 2.5 |
| P02647 | APOA1 | Apolipoprotein A-I | 28 | 2.2 × 10−5 | 1.3 | ˄ | 23.4 ± 0.7 | 21.6 ± 2.2 |
| P02652 | APOA2 | Apolipoprotein A-II | 6 | 1.2 × 10−6 | 3.3 | ˄ | 25.4 ± 0.9 | 23.6 ± 2.6 |
| P02671 | FGA | Fibrinogen alpha chain | 48 | 1.2 × 10−6 | 2.9 | ˄ | 23.4 ± 0.6 | 21.6 ± 2.1 |
| P02675 | FGB | Fibrinogen beta chain | 26 | 1.8 × 10−7 | 3.3 | ˄ | 23.6 ± 0.9 | 21.7 ± 2.5 |
| P02679 | FGG | Fibrinogen gamma chain | 28 | 2.4 × 10−7 | 3.1 | ˄ | 21.5 ± 0.9 | 19.6 ± 1.9 |
| P02747 | C1QC | Complement C1q subcomponent subunit C | 5 | 3.0 × 10−5 | 1.3 | ˄ | 27.6 ± 0.7 | 25.6 ± 2.1 |
| P02751 | FN1 | Fibronectin | 72 | 1.3 × 10−7 | 3.8 | ˄ | 24.9 ± 1 | 22.9 ± 2.3 |
| P02760 | AMBP | Protein AMBP | 5 | 1.2 × 10−6 | 1.4 | ˄ | 24.2 ± 0.9 | 22.1 ± 2.3 |
| P02787 | TF | Serotransferrin | 21 | 1.8 × 10−6 | 2.2 | ˄ | 25.1 ± 0.9 | 22.9 ± 2.8 |
| P04004 | VTN | Vitronectin | 15 | 6.5 × 10−6 | 2.1 | ˄ | 24.3 ± 1.1 | 22.1 ± 2.1 |
| P06727 | APOA4 | Apolipoprotein A-IV | 34 | 1.3 × 10−5 | 1.8 | ˄ | 24.2 ± 1.2 | 22 ± 2.6 |
| P07225 | PROS1 | Vitamin K-dependent protein S | 14 | 1.9 × 10−6 | 1.9 | ˄ | 22.4 ± 0.6 | 20.2 ± 2.4 |
| P08603 | CFH | Complement factor H | 18 | 2.0 × 10−6 | 2.1 | ˄ | 23.6 ± 1 | 21.2 ± 2.9 |
| P09871 | C1S | Complement C1s subcomponent | 17 | 4.5 × 10−7 | 2.6 | ˄ | 24.7 ± 0.8 | 22 ± 2.7 |
| P0C0L5 | C4B | Complement C4-B | 5 | 7.1 × 10−7 | 2.0 | ˄ | 25 ± 1.4 | 22.3 ± 2.6 |
| P10909 | CLU | Clusterin | 17 | 9.1 × 10−7 | 3.1 | ˄ | 28 ± 1.2 | 25.1 ± 3.1 |
| P12259 | F5 | Coagulation factor V | 39 | 1.4 × 10−5 | 1.6 | ˄ | 27.8 ± 1.2 | 24.6 ± 2.7 |
| P22352 | GPX3 | Glutathione peroxidase 3 | 6 | 2.3 × 10−6 | 1.9 | ˄ | 25.8 ± 1.1 | 22.7 ± 3.4 |
| P27169 | PON1 | Serum paraoxonase/arylesterase 1 | 10 | 5.0 × 10−6 | 2.2 | ˄ | 24.9 ± 1.4 | 21.6 ± 3.2 |
| P80108 | GPLD1 | Phosphatidylinositol-glycan-specific phospholipase D | 25 | 1.5 × 10−5 | 1.8 | ˄ | 28.2 ± 1 | 24.9 ± 3.1 |
| Q96KN2 | CNDP1 | Beta-Ala-His dipeptidase | 18 | 1.7 × 10−6 | 2.2 | ˄ | 26.7 ± 0.5 | 22.9 ± 3.7 |
| Protein IDs | Gene Names | Protein Names | Unique Peptides | p-Value | t-Test Difference | Log2 (LFQ Intensity) Background | Log2 (LFQ Intensity) + 1 Day | Log2 (LFQ Intensity) + 7 Days | |
|---|---|---|---|---|---|---|---|---|---|
| P02787 | TF | Serotransferrin | 26 | 1.70 × 10−7 | 2.5 | ˄ | 20.7 ± 2.1 | 23.2 ± 1.4 | 21.5 ± 1.9 |
| P04217 | A1BG | Alpha-1B-glycoprotein | 11 | 3.30 × 10−5 | 2.2 | ˄ | 17.1 ± 0.8 | 19.3 ± 1.1 | 18.3 ± 1.9 |
| P01008 | SERPINC1 | Antithrombin-III | 20 | 3.60 × 10−8 | 1.9 | ˄ | 19.1 ± 0.9 | 20.9 ± 1.2 | 19.6 ± 1.6 |
| P0DJI8 | SAA1 | Serum amyloid A-1 protein | 4 | 8.40 × 10−5 | 1.8 | ˄ | 19.6 ± 1.1 | 21.4 ± 1.4 | 20.2 ± 0.8 |
| P05155 | SERPING1 | Plasma protease C1 inhibitor | 11 | 1.70 × 10−4 | 1.8 | ˄ | 17.9 ± 1 | 19.7 ± 1.4 | 19.1 ± 1.4 |
| P00751 | CFB | Complement factor B | 13 | 9.50 × 10−6 | 1.8 | ˄ | 18.1 ± 1.3 | 19.9 ± 1.2 | 18.7 ± 1.5 |
| P01011 | SERPINA3 | Alpha-1-antichymotrypsin | 20 | 1.20 × 10−6 | 1.7 | ˄ | 19.1 ± 1.1 | 20.7 ± 1.1 | 20.3 ± 0.9 |
| P01023 | A2M | Alpha-2-macroglobulin | 40 | 5.60 × 10−5 | 1.4 | ˄ | 21.3 ± 1.8 | 22.8 ± 1.4 | 22.3 ± 1 |
| P01009 | SERPINA1 | Alpha-1-antitrypsin | 25 | 2.30 × 10−6 | 1.1 | ˄ | 21.2 ± 0.9 | 22.4 ± 1.1 | 21.9 ± 0.8 |
| P00738 | HP | Haptoglobin | 7 | 2.50 × 10−4 | 1 | ˄ | 21.1 ± 0.7 | 22.2 ± 1.4 | 21.3 ± 1.1 |
| P21333 | FLNA | Filamin-A | 11 | 2.20 × 10−4 | −0.6 | ˅ | 19.5 ± 0.2 | 18.9 ± 0.2 | 19.1 ± 0 |
| Q15166 | PON3 | Serum paraoxonase/lactonase 3 | 7 | 6.80 × 10−4 | −0.6 | ˅ | 19.6 ± 0.4 | 19 ± 0.7 | 19.4 ± 0.6 |
| O00391 | QSOX1 | Sulfhydryl oxidase 1 | 16 | 1.50 × 10−4 | −0.7 | ˅ | 20.5 ± 0.6 | 19.8 ± 0.8 | 20.3 ± 0.6 |
| P12259 | F5 | Coagulation factor V | 26 | 6.30 × 10−4 | −0.7 | ˅ | 21.1 ± 0.9 | 20.4 ± 0.8 | 20.9 ± 1 |
| P05160 | F13B | Coagulation factor XIII B chain | 8 | 3.30 × 10−4 | −0.8 | ˅ | 20.4 ± 0.9 | 19.6 ± 0.7 | 20 ± 0.9 |
| P33151 | CDH5 | Cadherin-5 | 8 | 4.70 × 10−4 | −0.8 | ˅ | 20 ± 0.4 | 19.2 ± 0.9 | 19.4 ± 0.7 |
| P00488 | F13A1 | Coagulation factor XIII A chain | 26 | 3.70 × 10−5 | −0.8 | ˅ | 21.3 ± 0.8 | 20.5 ± 0.8 | 21 ± 0.9 |
| P12830 | CDH1 | Cadherin-1 | 7 | 2.10 × 10−4 | −0.8 | ˅ | 18.8 ± 0.6 | 18 ± 0.4 | 18.8 ± 0.4 |
| Q12805 | EFEMP1 | EGF-containing fibulin-like extracellular matrix protein 1 | 5 | 4.40 × 10−6 | −1.2 | ˅ | 19.4 ± 0.5 | 18.1 ± 0.7 | 19 ± 0.7 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brzhozovskiy, A.G.; Kononikhin, A.S.; Pastushkova, L.C.; Kashirina, D.N.; Indeykina, M.I.; Popov, I.A.; Custaud, M.-A.; Larina, I.M.; Nikolaev, E.N. The Effects of Spaceflight Factors on the Human Plasma Proteome, Including Both Real Space Missions and Ground-Based Experiments. Int. J. Mol. Sci. 2019, 20, 3194. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20133194
Brzhozovskiy AG, Kononikhin AS, Pastushkova LC, Kashirina DN, Indeykina MI, Popov IA, Custaud M-A, Larina IM, Nikolaev EN. The Effects of Spaceflight Factors on the Human Plasma Proteome, Including Both Real Space Missions and Ground-Based Experiments. International Journal of Molecular Sciences. 2019; 20(13):3194. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20133194
Chicago/Turabian StyleBrzhozovskiy, Alexander G., Alexey S. Kononikhin, Lyudmila Ch. Pastushkova, Daria N. Kashirina, Maria I. Indeykina, Igor A. Popov, Marc-Antoine Custaud, Irina M. Larina, and Evgeny N. Nikolaev. 2019. "The Effects of Spaceflight Factors on the Human Plasma Proteome, Including Both Real Space Missions and Ground-Based Experiments" International Journal of Molecular Sciences 20, no. 13: 3194. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20133194