Testing Anti-Biofilm Polymeric Surfaces: Where to Start?
Abstract
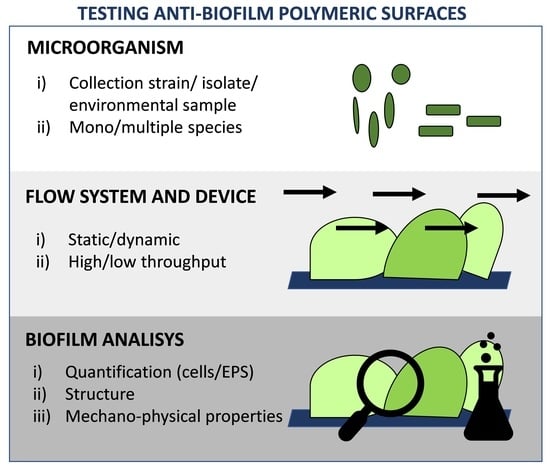
:1. Introduction
2. Anti-Biofilm Polymeric Surfaces
3. Microbial Choice
4. In Vitro Methods to Culture Biofilm on Anti-Biofilm Polymeric Surfaces
4.1. Static Methods
4.1.1. Microtiter Well Plate
4.1.2. Calgary Biofilm Device
4.1.3. The Biofilm Ring Test (BRT)
4.1.4. Real-Time xCelligence
4.1.5. Transwell Device
4.1.6. Colony Biofilm
4.2. Dynamic Methods
4.2.1. Robbins (RO) Reactor
4.2.2. Center for Disease Control (CDC) Reactor.
4.2.3. Rotating or Spinning Disk (RD) Reactor
4.2.4. Annular (AN) Reactor
4.2.5. Drip flow (DF) Reactor
4.2.6. Flow Cells (FC)
4.2.7. Microplate under Flow (Bioflux)
4.3. Microcosms
5. Quantitative Analysis of Biofilm on Anti-Biofilm Polymeric Surfaces
5.1. Viable Cellular Biomass
5.1.1. Plate Count Assay
5.1.2. Biomarker Quantification
Phospholipid Fatty Acids
Ergosterol
5.1.3. Metabolic Assays
Colorimetric Dyes
Adenosine Triphosphate (ATP) Bioluminescence
Isothermal Microcalorimetry (ICM)
Tunable Diode Laser Absorption Spectroscopy (TDLAS)
5.2. Total (Viable and Non-Viable) Cellular Biomass
5.2.1. Chamber Counting
5.2.2. Dye Binding
5.2.3. Biomarker Quantification
Total organic carbon
Proteins
Chlorophyll
5.2.4. Quantitative Polymerase Chain Reaction (qPCR)
5.2.5. Flow-Based Cell Counting
Coulter System
Flow Cytometry
5.3. Total Biofilm (Cellular + Extracellular Polymeric Substances (EPS) Components)
5.3.1. Dry Weight
5.3.2. Optical Density
5.3.3. Dye-Based Methods
5.3.4. Color Measurements
5.4. EPS Matrix
5.4.1. Ex Situ EPS Analysis
EPS Extraction
Proteins
Polysaccharides
Extracellular DNA
Antibody Microarrays
Spectroscopic Techniques
Advanced Techniques
5.4.2. In Situ EPS Analysis
Microscopic Techniques
Spectroscopic Techniques
6. Structural Assessment of Biofilm on Anti-Biofilm Polymeric Surfaces
6.1. Identification and Spatial Distribution of Biofilm Community Members
6.1.1. Fluorescence In Situ Hybridization (FISH)
6.1.2. Immunofluorescence Detection
6.2. Biofilm Imaging for Biofilm Morphology Studies
6.2.1. Confocal Laser Scanning Microscopy (CLSM)
6.2.2. Electron Microscopy
6.2.3. Super-Resolution Microscopy (Nanoscopy)
6.2.4. Other Microscopic Techniques
6.2.5. Optical Coherence Tomography (OCT)
6.3. Mechanical and Physical Properties
6.3.1. Atomic Force Microscopy (AFM)
6.3.2. Rheometry
6.3.3. Quartz Crystal Microbalance (QCM)
6.3.4. Surface Plasma Resonance (SPR)
6.3.5. Fluid Dynamic Gauging (FDG)
6.3.6. Microsensors
6.3.7. Magnetic Resonance Imaging (MRI) and Pulse-Field Gradient Nuclear Magnetic Resonance (PFG-NMR)
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Namazi, H. Polymers in our daily life. Bioimpacts 2017, 7, 73–74. [Google Scholar] [CrossRef]
- Teo, A.J.T.; Mishra, A.; Park, I.; Kim, Y.J.; Park, W.T.; Yoon, Y.J. Polymeric biomaterials for medical implants and devices. ACS Biomater. Sci. Eng. 2016, 2, 454–472. [Google Scholar] [CrossRef]
- Siracusa, V. Food packaging permeability behaviour: A report. Int. J. Polym. Sci. 2012, 2012, 302029. [Google Scholar] [CrossRef]
- Agrillo, B.; Balestrieri, M.; Gogliettino, M.; Palmieri, G.; Moretta, R.; Proroga, Y.T.; Rea, I.; Cornacchia, A.; Capuano, F.; Smaldone, G.; et al. Functionalized polymeric materials with bio-derived antimicrobial peptides for “active” packaging. Int. J. Mol. Sci. 2019, 20, 601. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.F.R.; Zhao, X.Y.; Sun, Y.; He, C.B.; Tan, M.C.; Tan, D.T.H. Low loss nanostructured polymers for chip-scale waveguide amplifiers. Sci. Rep. 2017, 7, 3366. [Google Scholar] [CrossRef] [PubMed]
- Cappitelli, F.; Zanardini, E.; Sorlini, C. The biodeterioration of synthetic resins used in conservation. Macromol. Biosci. 2004, 4, 399–406. [Google Scholar] [CrossRef]
- Giacomucci, L.; Toja, F.; Sanmartin, P.; Toniolo, L.; Prieto, B.; Villa, F.; Cappitelli, F. Degradation of nitrocellulose-based paint by Desulfovibrio desulfuricans ATCC 13541. Biodegradation 2012, 23, 705–716. [Google Scholar] [CrossRef] [PubMed]
- Haque, S.K.M.; Ardila-Rey, J.A.; Umar, Y.; Rahman, H.; Mas’ud, A.A.; Muhammad-Sukki, F.; Albarracin, R. Polymeric materials for conversion of electromagnetic waves from the sun to electric power. Polymers 2018, 10, 307. [Google Scholar] [CrossRef] [PubMed]
- Koniuszewska, A.G.; Kaczmar, J.W. Application of polymer based composite materials in transportation. Prog. Rubber Plast. Recycl. Technol. 2016, 32, 1–24. [Google Scholar] [CrossRef]
- Prabhu, T.N.; Prashantha, K.A. Review on present status and future challenges of starch based polymer films and their composites in food packaging applications. Polym. Compos. 2018, 39, 2499–2522. [Google Scholar] [CrossRef]
- Costerton, J.W. Introduction to biofilm. Int. J. Antimicrob. Agents 1999, 11, 217–221. [Google Scholar] [CrossRef]
- Plyuta, V.A.; Lipasova, V.A.; Kuznetsov, A.E.; Khmel, I.A. Effect of salicylic, indole-3-acetic, gibberellic, and abscisic acids on biofilm formation by Agrobacterium tumefaciens c58 and Pseudomonas aeruginosa PAO1. Appl. Biochem. Microbiol. 2013, 49, 706–710. [Google Scholar] [CrossRef]
- Riga, E.K.; Vohringer, M.; Widyaya, V.T.; Lienkamp, K. Polymer-based surfaces designed to reduce biofilm formation: From antimicrobial polymers to strategies for long-term applications. Macromol. Rapid Commun. 2017, 38. [Google Scholar] [CrossRef] [PubMed]
- Vigneron, A.; Alsop, E.B.; Chambers, B.; Lomans, B.P.; Head, I.M.; Tsesmetzis, N. Complementary microorganisms in highly corrosive biofilms from an offshore oil production facility. Appl. Environ. Microbiol. 2016, 82, 2545–2554. [Google Scholar] [CrossRef] [PubMed]
- Cappitelli, F.; Polo, A.; Villa, F. Biofilm formation in food processing environments is still poorly understood and controlled. Food Eng. Rev. 2014, 6, 29–42. [Google Scholar] [CrossRef]
- Douterelo, I.; Jackson, M.; Solomon, C.; Boxall, J. Spatial and temporal analogies in microbial communities in natural drinking water biofilms. Sci. Total Environ. 2017, 581, 277–288. [Google Scholar] [CrossRef] [PubMed]
- Galie, S.; Garcia-Gutierrez, C.; Miguelez, E.M.; Villar, C.J.; Lombo, F. Biofilms in the food industry: Health aspects and control methods. Front. Microbiol. 2018, 9, 898. [Google Scholar] [CrossRef]
- Francolini, I.; Donelli, G.; Crisante, F.; Taresco, V.; Piozzi, A. Antimicrobial polymers for anti-biofilm medical devices: State-of-art and perspectives. Adv. Exp. Med. Biol. 2015, 831, 93–117. [Google Scholar] [CrossRef]
- Sadekuzzaman, M.; Yang, S.; Mizan, M.F.R.; Ha, S.D. Current and recent advanced strategies for combating biofilms. Compr. Rev. Food Sci. Food Saf. 2015, 14, 491–509. [Google Scholar] [CrossRef]
- Anderson, D.J.; Podgorny, K.; Berrios-Torres, S.I.; Bratzler, D.W.; Dellinger, E.P.; Greene, L.; Nyquist, A.C.; Saiman, L.; Yokoe, D.S.; Maragakis, L.L.; et al. Strategies to prevent surgical site infections in acute care hospitals: 2014 update. Infect. Control Hosp. Epidemiol. 2014, 35, 605–627. [Google Scholar] [CrossRef]
- Badia, J.M.; Casey, A.L.; Petrosillo, N.; Hudson, P.M.; Mitchell, S.A.; Crosby, C. Impact of surgical site infection on healthcare costs and patient outcomes: A systematic review in six European countries. J. Hosp. Infect. 2017, 96, 1–15. [Google Scholar] [CrossRef]
- Lo, J.; Lange, D.; Chew, B.H. Ureteral stents and foley catheters-associated urinary tract infections: The role of coatings and materials in infection prevention. Antibiotics (Basel) 2014, 3, 87–97. [Google Scholar] [CrossRef]
- Johnson, J.R.; Kuskowski, M.A.; Wilt, T.J. Systematic review: Antimicrobial urinary catheters to prevent catheter-associated urinary tract infection in hospitalized patients. Ann. Intern. Med. 2006, 144, 116–126. [Google Scholar] [CrossRef]
- Stewart, P.S. Antimicrobial tolerance in biofilms. Microbiol. Spectr. 2015, 3. [Google Scholar] [CrossRef]
- Bas, S.; Kramer, M.; Stopar, D. Biofilm surface density determines biocide effectiveness. Front. Microbiol. 2017, 8, 2443. [Google Scholar] [CrossRef]
- Hoiby, N.; Bjarnsholt, T.; Givskov, M.; Molin, S.; Ciofu, O. Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents 2010, 35, 322–332. [Google Scholar] [CrossRef] [Green Version]
- Nisnevitch, M. Antibiotic resistance and antibiotic alternatives: Looking towards the future. Sci. Prog. 2016, 99, 92–96. [Google Scholar] [CrossRef]
- Alves, D.; Pereira, M.O. Bio-inspired coating strategies for the immobilization of polymyxins to generate contact-killing surfaces. Macromol. Biosci. 2016, 16, 1450–1460. [Google Scholar] [CrossRef]
- Sjollema, J.; Zaat, S.A.J.; Fontaine, V.; Ramstedt, M.; Luginbuehl, R.; Thevissen, K.; Li, J.Y.; van der Mei, H.C.; Busscher, H.J. In vitro methods for the evaluation of antimicrobial surface designs. Acta Biomater. 2018, 70, 12–24. [Google Scholar] [CrossRef]
- Phillips, K.S.; Patwardhan, D.; Jayan, G. Biofilms, medical devices, and antibiofilm technology: Key messages from a recent public workshop. Am. J. Infect. Control 2015, 43, 2–3. [Google Scholar] [CrossRef]
- Miquel, S.; Lagrafeuille, R.; Souweine, B.; Forestier, C. Anti-biofilm activity as a health issue. Front. Microbiol. 2016, 7, 592. [Google Scholar] [CrossRef] [PubMed]
- Roy, R.; Tiwari, M.; Donelli, G.; Tiwari, V. Strategies for combating bacterial biofilms: A focus on anti-biofilm agents and their mechanisms of action. Virulence 2018, 9, 522–554. [Google Scholar] [CrossRef] [PubMed]
- Yu, O.Y.; Zhao, I.S.; Mei, M.L.; Lo, E.C.; Chu, C.H. Dental biofilm and laboratory microbial culture models for cariology research. Dent. J. (Basel) 2017, 5, 21. [Google Scholar] [CrossRef]
- Haney, E.F.; Trimble, M.J.; Cheng, J.T.; Valle, Q.; Hancock, R.E.W. Critical assessment of methods to quantify biofilm growth and evaluate antibiofilm activity of host defence peptides. Biomolecules 2018, 8, 29. [Google Scholar] [CrossRef] [PubMed]
- Azeredo, J.; Azevedo, N.F.; Briandet, R.; Cerca, N.; Coenye, T.; Costa, A.R.; Desvaux, M.; Di Bonaventura, G.; Hébraud, M.; Jaglic, Z.; et al. Critical review on biofilm methods. Crit. Rev. Microbiol. 2017, 43, 313–351. [Google Scholar] [CrossRef] [PubMed]
- Cattò, C.; Villa, F.; Cappitelli, F. Recent progress in bio-inspired biofilm-resistant polymeric surfaces. Crit. Rev. Microbiol. 2018, 44, 633–652. [Google Scholar] [CrossRef] [PubMed]
- Lichter, J.A.; Van Vliet, K.J.; Rubner, M.F. Design of antibacterial surfaces and interfaces: Polyelectrolyte multilayers as a multifunctional platform. Macromolecules 2009, 42, 8573–8586. [Google Scholar] [CrossRef]
- Ganewatta, M.S.; Miller, K.P.; Singleton, S.P.; Mehrpouya-Bahrami, P.; Chen, Y.P.; Yan, Y.; Nagarkatti, M.; Nagarkatti, P.; Decho, A.W.; Tang, C. Antibacterial and biofilm-disrupting coatings from resin acid-derived materials. Biomacromolecules 2015, 16, 3336–3344. [Google Scholar] [CrossRef]
- Antoci, V.; Adams, C.S.; Parvizi, J.; Davidson, H.M.; Composto, R.J.; Freeman, T.A.; Wickstrom, E.; Ducheyne, P.; Jungkind, D.; Shapiro, I.M.; et al. The inhibition of Staphylococcus epidermidis biofilm formation by vancomycin-modified titanium alloy and implications for the treatment of periprosthetic infection. Biomaterials 2008, 29, 4684–4690. [Google Scholar] [CrossRef]
- Francolini, I.; Donelli, G. Prevention and control of biofilm-based medical-device-related infections. FEMS Immunol. Med. Microbiol. 2010, 59, 227–238. [Google Scholar] [CrossRef] [Green Version]
- Swartjes, J.; Sharma, P.K.; van Kooten, T.G.; van der Mei, H.C.; Mahmoudi, M.; Busscher, H.J.; Rochford, E.T.J. Current developments in antimicrobial surface coatings for biomedical applications. Curr. Med. Chem. 2015, 22, 2116–2129. [Google Scholar] [CrossRef] [PubMed]
- Ashbaugh, A.G.; Jiang, X.S.; Zheng, J.; Tsai, A.S.; Kim, W.S.; Thompson, J.M.; Miller, R.J.; Shahbazian, J.H.; Wang, Y.; Dillen, C.A.; et al. Polymeric nanofiber coating with tunable combinatorial antibiotic delivery prevents biofilm-associated infection in vivo. Proc. Natl. Acad. Sci. USA 2016, 113, E6919–E6928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barde, M.; Davis, M.; Rangari, S.; Mendis, H.C.; De La Fuente, L.; Auad, M.L. Development of antimicrobial-loaded polyurethane films for drug-eluting catheters. J. Appl. Polym. Sci. 2018, 135, 46467. [Google Scholar] [CrossRef]
- Coenye, T.; Nelis, H.J. In vitro and in vivo model systems to study microbial biofilm formation. J. Microbiol. Methods 2010, 83, 89–105. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Nie, X.; Zou, M.J.; Shi, Y.J.; Cheng, G. Recent advances in materials for extended-release antibiotic delivery system. J. Antibiot. 2011, 64, 625–634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, M.; Yu, Q.S.; Sun, H.M. Novel strategies for the prevention and treatment of biofilm related infections. Int. J. Mol. Sci. 2013, 14, 18488–18501. [Google Scholar] [CrossRef] [PubMed]
- Zanini, S.; Polissi, A.; Maccagni, E.A.; Dell’Orto, E.C.; Liberatore, C.; Riccardi, C. Development of antibacterial quaternary ammonium silane coatings on polyurethane catheters. J. Colloid Interface Sci. 2015, 451, 78–84. [Google Scholar] [CrossRef] [PubMed]
- Ergene, C.; Palermo, E.F. Cationic poly (benzyl ether) s as self-Immolative antimicrobial polymers. Biomacromolecules 2017, 18, 3400–3409. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wu, B.; Chen, H.; Nan, K.H.; Jin, Y.Y.; Sun, L.; Wang, B.L. Recent developments in smart antibacterial surfaces to inhibit biofilm formation and bacterial infections. J. Mater. Chem. B 2018, 6, 4274–4292. [Google Scholar] [CrossRef]
- Gbejuade, H.O.; Lovering, A.M.; Webb, J.C. The role of microbial biofilms in prosthetic joint infections. Acta Orthop. 2015, 86, 147–158. [Google Scholar] [CrossRef]
- Romano, C.L.; Scarponi, S.; Gallazzi, E.; Romano, D.; Drago, L. Antibacterial coating of implants in orthopaedics and trauma: A classification proposal in an evolving panorama. J. Orthop. Surg. Res. 2015, 10, 157. [Google Scholar] [CrossRef] [PubMed]
- Adlhart, C.; Verran, J.; Azevedo, N.F.; Olmez, H.; Keinanen-Toivola, M.M.; Gouveia, I.; Melo, L.F.; Crijns, F. Surface modifications for antimicrobial effects in the healthcare setting: A critical overview. J. Hosp. Infect. 2018, 99, 239–249. [Google Scholar] [CrossRef] [PubMed]
- Ding, W.; Ma, C.F.; Zhang, W.P.; Chiang, H.Y.; Tam, C.; Xu, Y.; Zhang, G.Z.; Qian, P.Y. Anti-biofilm effect of a butenolide/polymer coating and metatranscriptomic analyses. Biofouling 2018, 34, 111–122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Francolini, I.; Silvestro, I.; Di Lisio, V.; Martinelli, A.; Piozzi, A. Synthesis, characterization, and bacterial fouling-resistance properties of polyethylene glycol-grafted polyurethane elastomers. Int. J. Mol. Sci. 2019, 20, 1001. [Google Scholar] [CrossRef] [PubMed]
- Hsu, L.C.; Fang, J.; Borca-Tasciuc, D.A.; Worobo, R.W.; Moraru, C.I. Effect of micro- and nanoscale topography on the adhesion of bacterial cells to solid surfaces. Appl. Environ. Microbiol. 2013, 79, 2703–2712. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, I.C.S.; van der Mei, H.C.; Lochhead, M.J.; Grainger, D.W.; Busscher, H.J. The inhibition of the adhesion of clinically isolated bacterial strains on multi-component cross-linked poly(ethylene glycol)-based polymer coatings. Biomaterials 2007, 28, 4105–4112. [Google Scholar] [CrossRef] [PubMed]
- Epstein, A.K.; Wong, T.S.; Belisle, R.A.; Boggs, E.M.; Aizenberg, J. Liquid-infused structured surfaces with exceptional anti-biofouling performance. Proc. Natl. Acad. Sci. USA 2012, 109, 13182–13187. [Google Scholar] [CrossRef] [Green Version]
- Zaltsman, N.; Ionescu, A.C.; Weiss, E.I.; Brambilla, E.; Beyth, S.; Beyth, N. Surface-modified nanoparticles as anti-biofilm filler for dental polymers. PLoS ONE 2017, 12, e0189397. [Google Scholar] [CrossRef]
- Cao, W.W.; Zhang, Y.; Wang, X.; Li, Q.; Xiao, Y.H.; Li, P.L.; Wang, L.N.; Ye, Z.W.; Xing, X.D. Novel resin-based dental material with anti-biofilm activity and improved mechanical property by incorporating hydrophilic cationic copolymer functionalized nanodiamond. J. Mater. Sci. Mater. Med. 2018, 29, 162. [Google Scholar] [CrossRef]
- Knowles, B.R.; Wagner, P.; Maclaughlin, S.; Higgins, M.J.; Molino, P.J. Silica nanoparticles functionalized with zwitterionic sulfobetaine siloxane for application as a versatile antifouling coating system. ACS Appl. Mater. Interfaces 2017, 9, 18584–18594. [Google Scholar] [CrossRef]
- Gkana, E.N.; Doulgeraki, A.I.; Chorianopoulos, N.G.; Nychas, G.J.E. Anti-adhesion and anti-biofilm potential of organosilane nanoparticles against foodborne pathogens. Front. Microbiol. 2017, 8, 1295. [Google Scholar] [CrossRef] [PubMed]
- Sufian, M.M.; Khattak, J.Z.K.; Yousaf, S.; Rana, M.S. Safety issues associated with the use of nanoparticles in human body. Photodiagnosis Photodyn. Ther. 2017, 19, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Reed, R.B.; Zaikova, T.; Barber, A.; Simonich, M.; Lankone, R.; Marco, M.; Hristovski, K.; Herckes, P.; Passantino, L.; Fairbrother, D.H.; et al. Potential environmental impacts and antimicrobial efficacy of silver and nanosilver-containing textiles. Environ. Sci. Technol. 2016, 50, 4018–4026. [Google Scholar] [CrossRef] [PubMed]
- Resnik, D.B. How should engineered nanomaterials be regulated for public and environmental health? AMA J. Ethics 2019, 21, 363–369. [Google Scholar] [CrossRef]
- Villa, F.; Cappitelli, F. Plant-derived bioactive compounds at sub-lethal concentrations: Towards smart biocide-free antibiofilm strategies. Phytochem. Rev. 2013, 12, 245–254. [Google Scholar] [CrossRef]
- Rémy, B.; Mion, S.; Plener, L.; Elias, M.; Chabrière, E.; Daudé, D. Interference in bacterial quorum sensing: A biopharmaceutical perspective. Front. Pharmacol. 2018, 9, 203. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, J.B. Biofilm dispersal: Mechanisms, clinical implications, and potential therapeutic uses. J. Dent. Res. 2010, 89, 205–218. [Google Scholar] [CrossRef] [PubMed]
- Kostakioti, M.; Hadjifrangiskou, M.; Hultgren, S.J. Bacterial biofilms: Development, dispersal, and therapeutic strategies in the dawn of the postantibiotic era. Cold Spring Harb. Perspect. Med. 2013, 3, a010306. [Google Scholar] [CrossRef] [PubMed]
- Chifiriuc, C.; Grumezescu, V.; Grumezescu, A.M.; Saviuc, C.; Lazăr, V.; Andronescu, E. Hybrid magnetite nanoparticles/Rosmarinus officinalis essential oil nanobiosystem with antibiofilm activity. Nanoscale Res. Lett. 2012, 7, 209. [Google Scholar] [CrossRef] [PubMed]
- Nowatzki, P.J.; Koepsel, R.R.; Stoodley, P.; Min, K.; Harper, A.; Murata, H.; Donfack, J.; Hortelano, E.R.; Ehrlich, G.D.; Russell, A.J. Salicylic acid-releasing polyurethane acrylate polymers as anti-biofilm urological catheter coatings. Acta Biomater. 2012, 8, 1869–1880. [Google Scholar] [CrossRef]
- Marcano, A.; Ba, O.; Thebault, P.; Crétois, R.; Marais, S.; Duncan, A.C. Elucidation of innovative antibiofilm materials. Colloids Surf. B 2015, 136, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Villa, F.; Secundo, F.; Polo, A.; Cappitelli, F. Immobilized hydrolytic enzymes exhibit antibiofilm activity against Escherichia coli at sub-lethal concentrations. Curr. Microbiol. 2015, 71, 106–114. [Google Scholar] [CrossRef] [PubMed]
- Spadoni-Andreani, E.; Villa, F.; Cappitelli, F.; Krasowska, A.; Biniarz, P.; Lukaszewicz, M.; Secundo, F. Coating polypropylene surfaces with protease weakens the adhesion and increases the dispersion of Candida albicans cells. Biotechnol. Lett. 2017, 39, 423–428. [Google Scholar] [CrossRef] [PubMed]
- Cattò, C.; Secundo, F.; James, G.; Villa, F.; Cappitelli, F. α-Chymotrypsin immobilized on a low-density polyethylene surface successfully weakens Escherichia coli biofilm formation. Int. J. Mol. Sci. 2018, 19, 4003. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.G.; Lee, J.H.; Gwon, G.; Kim, S.I.; Park, J.G.; Lee, J. Essential oils and eugenols inhibit biofilm formation and the virulence of Escherichia coli O157:H7. Sci. Rep. 2016, 6, 36377. [Google Scholar] [CrossRef] [PubMed]
- Dell’orto, S.; Catto, C.; Villa, F.; Forlani, F.; Vassallo, E.; Morra, M.; Cappitelli, F.; Villa, S.; Gelain, A. Low density polyethylene functionalized with antibiofilm compounds inhibits Escherichia coli cell adhesion. J. Biomed. Mater. Res. A 2017, 105, 3251–3261. [Google Scholar] [CrossRef] [PubMed]
- Sajeevan, S.E.; Chatterjee, M.; Paul, V.; Baranwal, G.; Kumar, V.A.; Bose, C.; Banerji, A.; Nair, B.G.; Prasanth, B.P.; Biswas, R. Impregnation of catheters with anacardic acid from cashew nut shell prevents Staphylococcus aureus biofilm development. J. Appl. Microbiol. 2018, 125, 1286–1295. [Google Scholar] [CrossRef]
- Pu, Y.; Liu, A.B.; Zheng, Y.Q.; Ye, B. In vitro damage of Candida albicans biofilms by chitosan. Exp. Ther. Med. 2014, 8, 929–934. [Google Scholar] [CrossRef]
- Bregnocchi, A.; Zanni, E.; Uccelletti, D.; Marra, F.; Cavallini, D.; De Angelis, F.; De Bellis, G.; Bossu, M.; Ierardo, G.; Polimeni, A.; et al. Graphene-based dental adhesive with anti-biofilm activity. J. Nanobiotechnol. 2017, 15, 89. [Google Scholar] [CrossRef]
- Namivandi-Zangeneh, R.; Sadrearhami, Z.; Bagheri, A.; Sauvage-Nguyen, M.; Ho, K.K.K.; Kumar, N.; Wong, E.H.H.; Boyer, C. Nitric oxide-loaded antimicrobial polymer for the synergistic eradication of bacterial biofilm. ACS Macro Lett. 2018, 7, 592–597. [Google Scholar] [CrossRef]
- Le Norcy, T.; Niemann, H.; Proksch, P.; Linossier, I.; Vallee-Rehel, K.; Hellio, C.; Fay, F. Anti-biofilm effect of biodegradable coatings based on hemibastadin derivative in marine environment. Int. J. Mol. Sci. 2017, 18, E1520. [Google Scholar] [CrossRef]
- Liu, H.Y.; Shukla, S.; Vera-Gonzalez, N.; Tharmalingam, N.; Mylonakis, E.; Fuchs, B.B.; Shukla, A. Auranofin releasing antibacterial and antibiofilm polyurethane intravascular catheter coatings. Front. Cell. Infect. Microbiol. 2019, 9, 37. [Google Scholar] [CrossRef]
- Zhang, N.; Zhang, K.; Melo, M.A.S.; Weir, M.D.; Xu, D.J.; Bai, Y.X.; Xu, H.H.K. Effects of long-term water-aging on novel anti-biofilm and protein-repellent dental composite. Int. J. Mol. Sci. 2017, 18, 186. [Google Scholar] [CrossRef]
- Perez-Nadales, E.; Nogueira, M.F.A.; Baldin, C.; Castanheira, S.; El Ghalid, M.; Grund, E.; Lengeler, K.; Marchegiani, E.; Mehrotra, P.V.; Moretti, M.; et al. Fungal model systems and the elucidation of pathogenicity determinants. Fungal Genet. Biol. 2014, 70, 42–67. [Google Scholar] [CrossRef] [Green Version]
- Nett, J.E.; Andes, D.R. Fungal Biofilms: In vivo models for discovery of anti-biofilm drugs. Microbiol. Spectr. 2015, 3, E30. [Google Scholar] [CrossRef]
- Villa, F.; Pitts, B.; Lauchnor, E.; Cappitelli, F.; Stewart, P.S. Development of a laboratory model of a phototroph-heterotroph mixed-species biofilm at the stone/air interface. Front. Microbiol. 2015, 6, 1251. [Google Scholar] [CrossRef]
- Peng, C.; Vishwakarma, A.; Li, Z.R.; Miyoshi, T.; Barton, H.A.; Joy, A. Modification of a conventional polyurethane composition provides significant anti-biofilm activity against Escherichia coli. Polym. Chem. 2018, 9, 3195–3198. [Google Scholar] [CrossRef]
- Karathia, H.; Vilaprinyo, E.; Sorribas, A.; Alves, R. Saccharomyces cerevisiae as a Model Organism: A Comparative Study. PLoS ONE 2011, 6, e16015. [Google Scholar] [CrossRef]
- Russell, J.J.; Theriot, J.A.; Sood, P.; Marshall, W.F.; Landweber, L.F.; Fritz-Laylin, L.; Polka, J.K.; Oliferenko, S.; Gerbich, T.; Gladfelter, A.; et al. Non-model model organisms. BMC Biol. 2017, 15, 55. [Google Scholar] [CrossRef]
- Lebeaux, D.; Chauhan, A.; Rendueles, O.; Beloin, C. From in vitro to in vivo models of bacterial biofilm-related infections. Pathogens 2013, 2, 288–356. [Google Scholar] [CrossRef]
- Roder, H.L.; Sorensen, S.J.; Burmolle, M. Studying Bacterial multispecies biofilms: Where to start? Trends Microbiol. 2016, 24, 503–513. [Google Scholar] [CrossRef]
- Rzhepishevska, O.; Limanska, N.; Galkin, M.; Lacoma, A.; Lundquist, M.; Sokol, D.; Hakobyan, S.; Sjostedt, A.; Prat, C.; Ramstedt, M. Characterization of clinically relevant model bacterial strains of Pseudomonas aeruginosa for anti-biofilm testing of materials. Acta Biomater. 2018, 76, 99–107. [Google Scholar] [CrossRef]
- Eydallin, G.; Ryall, B.; Maharjan, R.; Ferenci, T. The nature of laboratory domestication changes in freshly isolated Escherichia coli strains. Environ. Microbiol. 2014, 16, 813–828. [Google Scholar] [CrossRef]
- Elias, S.; Banin, E. Multi-species biofilms: Living with friendly neighbors. FEMS Microbiol. Rev. 2012, 36, 990–1004. [Google Scholar] [CrossRef]
- Liu, W.Z.; Roder, H.L.; Madsen, J.S.; Bjarnsholt, T.; Sorensen, S.J.; Burmolle, M. Interspecific bacterial interactions are reflected in multispecies biofilm spatial organization. Front. Microbiol. 2016, 7, 1366. [Google Scholar] [CrossRef]
- Tay, W.H.; Chong, K.K.L.; Kline, K.A. Polymicrobial-host interactions during infection. J. Mol. Biol. 2016, 428, 3355–3371. [Google Scholar] [CrossRef]
- Parijs, I.; Steenackers, H.P. Competitive inter-species interactions underlie the increased antimicrobial tolerance in multispecies brewery biofilms. ISME J. 2018, 12, 2061–2075. [Google Scholar] [CrossRef] [Green Version]
- Tan, C.H.; Lee, K.W.K.; Burmolle, M.; Kjelleberg, S.; Rice, S.A. All together now: Experimental multispecies biofilm model systems. Environ. Microbiol. 2017, 19, 42–53. [Google Scholar] [CrossRef]
- Zhou, C.; Wu, Y.; Thappeta, K.R.V.; Subramanian, J.T.L.; Pranantyo, D.; Kang, E.T.; Duan, H.W.; Kline, K.; Chan-Park, M.B. In vivo anti-biofilm and anti-bacterial non-leachable coating thermally polymerized on cylindrical catheter. ACS Appl. Mater. Interfaces 2017, 9, 36269–36280. [Google Scholar] [CrossRef]
- Albuquerque, M.T.P.; Nagata, J.; Bottino, M.C. Antimicrobial efficacy of triple antibiotic-eluting polymer nanofibers against multispecies biofilm. Acta Biomater. 2017, 43, S51–S56. [Google Scholar] [CrossRef]
- Cossu, A.; Si, Y.; Sun, G.; Nitin, N. Antibiofilm effect of poly(vinyl alcohol-co-ethylene) halamine film against Listeria innocua and Escherichia coli O157:H7. Appl. Environ. Microbiol. 2017, 83, e00975-17. [Google Scholar] [CrossRef]
- Kommerein, N.; Stumpp, S.N.; Musken, M.; Ehlert, N.; Winkel, A.; Haussler, S.; Behrens, P.; Buettner, F.F.R.; Stiesch, M. An oral multispecies biofilm model for high content screening applications. PLoS ONE 2017, 12, e0173973. [Google Scholar] [CrossRef]
- Roeselers, G.; Zippel, B.; Staal, M.; van Loosdrecht, M.; Muyzer, G. On the reproducibility of microcosm experiments-different community composition in parallel phototrophic biofilm microcosms. FEMS Microbiol. Ecol. 2006, 58, 169–178. [Google Scholar] [CrossRef]
- American Society for Testing and Materials. ASTM E2799-17. Standard Test Method for Testing Disinfectant Efficacy Against Pseudomonas Aeruginosa Biofilm Using the MBEC Assay; ASTM International: West Conshohocken, PA, USA, 2017. [Google Scholar]
- American Society for Testing and Materials. ASTM E2562-17. Standard Test Method for Quantification of Pseudomonas Aeruginosa Biofilm Grown with High Shear and Continuous Flow Using CDC Biofilm Reactor; ASTM International: West Conshohocken, PA, USA, 2017. [Google Scholar]
- American Society for Testing and Materials. ASTM E2871-19. Standard Test Method for Determining Disinfectant Efficacy Against Biofilm Grown in the CDC Biofilm Reactor Using the Single Tube Method; ASTM International: West Conshohocken, PA, USA, 2019. [Google Scholar]
- American Society for Testing and Materials. ASTM E2196-17. Standard Test Method for Quantification of Pseudomonas Aeruginosa Biofilm Grown with Medium Shear and Continuous Flow Using Rotating Disk Reactor; ASTM International: West Conshohocken, PA, USA, 2017. [Google Scholar]
- American Society for Testing and Materials. ASTM E2647-13. Standard Test Method for Quantification of Pseudomonas Aeruginosa Biofilm Grown Using Drip Flow Biofilm Reactor with Low Shear and Continuous Flow; ASTM International: West Conshohocken, PA, USA, 2013. [Google Scholar]
- Merritt, J.H.; Kadouri, D.E.; O’Toole, G.A. Growing and analyzing static biofilms. Curr. Protoc. Microbiol. 2005, 1, 1B.1.1–1B.1.17. [Google Scholar] [CrossRef]
- O’Toole, G.A. Microtiter dish biofilm formation assay. J. Vis. Exp. 2011, 47, 2437. [Google Scholar] [CrossRef]
- Lin, M.H.; Chang, F.R.; Hua, M.Y.; Wu, Y.C.; Liu, S.T. Inhibitory effects of 1,2,3,4,6-penta-O-galloyl-beta-D-glucopyranose on biofilm formation by Staphylococcus aureus. Antimicrob. Agents Chemother. 2011, 55, 1021–1027. [Google Scholar] [CrossRef]
- Swartjes, J.; Das, T.; Sharifi, S.; Subbiahdoss, G.; Sharma, P.K.; Krom, B.P.; Busscher, H.J.; van der Mei, H.C. A Functional DNase I coating to prevent adhesion of bacteria and the formation of biofilm. Adv. Funct. Mater. 2013, 23, 2843–2849. [Google Scholar] [CrossRef]
- Salta, M.; Dennington, S.P.; Wharton, J.A. Biofilm inhibition by novel natural product- and biocide-containing coatings using high-throughput screening. Int. J. Mol. Sci. 2018, 19, 1434. [Google Scholar] [CrossRef]
- Ceri, H.; Olson, M.E.; Stremick, C.; Read, R.R.; Morck, D.; Buret, A. The Calgary biofilm device: New technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J. Clin. Microbiol. Infect. 1999, 37, 1771–1776. [Google Scholar]
- Franklin, M.J.; Chang, C.; Akiyama, T.; Bothner, B. New Technologies for studying biofilms. Microbiol. Spectr. 2015, 3. [Google Scholar] [CrossRef]
- Harrison, J.J.; Ceri, H.; Yerly, J.; Stremick, C.A.; Hu, Y.P.; Martinuzzi, R.; Turner, R.J. The use of microscopy and three-dimensional visualization to evaluate the structure of microbial biofilms cultivated in the Calgary Biofilm Device. Biol. Proced. Online 2006, 8, 194–215. [Google Scholar] [CrossRef] [PubMed]
- Olivares, E.; Badel-Berchoux, S.; Provot, C.; Jaulhac, B.; Prevost, G.; Bernardi, T.; Jehl, F. The biofilm ring test: A rapid method for routine analysis of Pseudomonas aeruginosa biofilm formation kinetics. J. Clin. Microbiol. 2016, 54, 657–661. [Google Scholar] [CrossRef] [PubMed]
- Magana, M.; Sereti, C.; Ioannidis, A.; Mitchell, C.A.; Ball, A.R.; Magiorkinis, E.; Chatzipanagiotou, S.; Hamblin, M.R.; Hadjifrangiskou, M.; Tegos, G.P. Options and limitations in clinical investigation of bacterial biofilms. Clin. Microbiol. Rev. 2018, 31, e00084-16. [Google Scholar] [CrossRef] [PubMed]
- Stadelmaier, H.H. Magnetic properties of materials. Mater Sci. Eng. A Struct. Mater. 2000, 287, 138–145. [Google Scholar] [CrossRef]
- Junka, A.F.; Janczura, A.; Smutnicka, D.; Maczynska, B.; Secewicz, A.; Nowicka, J.; Bartoszewicz, M.; Gosciniak, G. Use of the real time xCelligence system for purposes of medical microbiology. Pol. J. Microbiol. 2012, 61, 191–197. [Google Scholar] [PubMed]
- Gutierrez, D.; Hidalgo-Cantabrana, C.; Rodriguez, A.; Garcia, P.; Ruas-Madiedo, P. Monitoring in real time the formation and removal of biofilms from clinical related pathogens using an impedance-based technology. PLoS ONE 2016, 11, e0163966. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, M.D.; Rodriguez, J.C.; Alvarez, L.; Artacho, A.; Royo, G.; Mira, A. Effect of antibiotics on biofilm inhibition and induction measured by real-time cell analysis. J. Appl. Microbiol. 2017, 122, 640–650. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, D.; Fernandez, L.; Martinez, B.; Ruas-Madiedo, P.; Garcia, P.; Rodriguez, A. Real-Time assessment of Staphylococcus aureus biofilm disruption by phage-derived proteins. Front. Microbiol. 2017, 8, 1632. [Google Scholar] [CrossRef]
- Aggas, J.R.; Harrell, W.; Lutkenhaus, J.; Guiseppi-Elie, A. Metal-polymer interface influences apparent electrical properties of nano-structured polyaniline films. Nanoscale 2018, 10, 672–682. [Google Scholar] [CrossRef]
- Wu, C.C.; Lin, C.T.; Wu, C.Y.; Peng, W.S.; Lee, M.J.; Tsai, Y.C. Inhibitory effect of Lactobacillus salivarius on Streptococcus mutans biofilm formation. Mol. Oral Microbiol. 2015, 30, 16–26. [Google Scholar] [CrossRef]
- Wang, Z.L.; Xiang, Q.Q.; Yang, T.; Li, L.Q.; Yang, J.L.; Li, H.G.; He, Y.; Zhang, Y.H.; Lu, Q.; Yu, J.L. Autoinducer-2 of Streptococcus mitis as a target molecule to inhibit pathogenic multi-species biofilm formation in vitro and in an endotracheal intubation rat model. Front. Microbiol. 2016, 7, 88. [Google Scholar] [CrossRef] [PubMed]
- Powell, L.C.; Pritchard, M.F.; Ferguson, E.L.; Powell, K.A.; Patel, S.U.; Rye, P.D.; Sakellakou, S.M.; Buurma, N.J.; Brilliant, C.D.; Copping, J.M.; et al. Targeted disruption of the extracellular polymeric network of Pseudomonas aeruginosa biofilms by alginate oligosaccharides. Npj Biofilms Microbiomes 2018, 4, 13. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.Y.; Thyiam, G.; Kang, J.E.; Lee, S.H.; Park, S.H.; Kim, J.S.; Abraham, M. Development of an Escherichia coli biofilm model on transwell. Korean J. Clin. Lab. Sci. 2012, 44, 112–117. [Google Scholar] [CrossRef]
- Standar, K.; Kreikemeyer, B.; Redanz, S.; Munter, W.L.; Laue, M.; Podbielski, A. Setup of an in vitro test system for basic studies on biofilm behavior of mixed-species cultures with dental and periodontal pathogens. Plos ONE 2010, 5, e13135. [Google Scholar] [CrossRef] [PubMed]
- Peterson, S.B.; Irie, Y.; Borlee, B.R.; Murakami, K.; Harrison, J.J.; Colvin, K.M.; Parsek, M.R. Different Methods for culturing biofilms in vitro. In Biofilm Infections; Bjarnsholt, T., Jensen, P., Moser, C., Høiby, N., Eds.; Springer: New York, NY, USA, 2011; pp. 251–266. ISBN 978-1-4419-6084-9. [Google Scholar]
- Tran, P.L.; Hammond, A.A.; Mosley, T.; Cortez, J.; Gray, T.; Colmer-Hamood, J.A.; Shashtri, M.; Spallholz, J.E.; Hamood, A.N.; Reid, T.W. Organoselenium coating on cellulose inhibits the formation of biofilms by Pseudomonas aeruginosa and Staphylococcus aureus. Appl. Environ. Microbiol. 2009, 75, 3586–3592. [Google Scholar] [CrossRef] [PubMed]
- Bakker, D.P.; van der Mats, A.; Verkerke, G.J.; Busscher, H.J.; van der Mei, H.C. Comparison of velocity profiles for different flow chamber designs used in studies of microbial adhesion to surfaces. Appl. Environ. Microbiol. 2003, 69, 6280–6287. [Google Scholar] [CrossRef]
- Goeres, D.M.; Loetterle, L.R.; Hamilton, M.A.; Murga, R.; Kirby, D.W.; Donlan, R.M. Statistical assessment of a laboratory method for growing biofilms. Microbiology 2005, 151, 757–762. [Google Scholar] [CrossRef] [Green Version]
- Gomes, I.B.; Meireles, A.; Goncalves, A.L.; Goeres, D.M.; Sjollema, J.; Simoes, L.C.; Simoes, M. Standardized reactors for the study of medical biofilms: A review of the principles and latest modifications. Crit. Rev. Biotechnol. 2018, 38, 657–670. [Google Scholar] [CrossRef]
- Schwartz, K.; Stephenson, R.; Hernandez, M.; Jambang, N.; Boles, B.R. The use of drip flow and rotating disk reactors for Staphylococcus aureus biofilm analysis. J. Vis. Exp. 2010, 46, 2470. [Google Scholar] [CrossRef]
- Sebestyen, P.; Blanken, W.; Bacsa, I.; Toth, G.; Martin, A.; Bhaiji, T.; Dergez, A.; Kesseru, P.; Koos, A.; Kiss, I. Upscale of a laboratory rotating disk biofilm reactor and evaluation of its performance over a half-year operation period in outdoor conditions. Algal Res. 2016, 18, 266–272. [Google Scholar] [CrossRef]
- Linton, C.J.; Sherriff, A.; Millar, M.R. Use of a modified Robbins device to directly compare the adhesion of Staphylococcus epidermidis RP62A to surfaces. J. Appl. Microbiol. 1999, 86, 194–202. [Google Scholar] [CrossRef] [PubMed]
- McCoy, W.F.; Bryers, J.D.; Robbins, J.; Costerton, J.W. Observations of fouling biofilm formation. Can. J. Microbiol. 1981, 27, 910–917. [Google Scholar] [CrossRef] [PubMed]
- Oosterhof, J.J.H.; Buijssen, K.; Busscher, H.J.; van der Laan, B.; van der Mei, H.C. Effects of quaternary ammonium silane coatings on mixed fungal and bacterial biofilms on tracheoesophageal shunt prostheses. Appl. Environ. Microbiol. 2006, 72, 3673–3677. [Google Scholar] [CrossRef] [PubMed]
- Ramage, G.; Wickes, B.L.; Lopez-Ribot, J.L. A seed and feed model for the formation of Candida albicans biofilms under flow conditions using an improved modified Robbins device. Rev. Iberoam. Micol. 2008, 25, 37–40. [Google Scholar] [CrossRef]
- Ginige, M.P.; Garbin, S.; Wylie, J.; Krishna, K.C.B. Effectiveness of devices to monitor biofouling and metals deposition on plumbing materials exposed to a full-scale drinking water distribution system. PLoS ONE 2017, 12, e0169140. [Google Scholar] [CrossRef] [PubMed]
- Cai, W.Y.; Wu, J.F.; Xi, C.W.; Meyerhoff, M.E. Diazeniumdiolate-doped poly(lactic-co-glycolic acid)-based nitric oxide releasing films as antibiofilm coatings. Biomaterials 2012, 33, 7933–7944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Carrera, C.; Chen, R.; Li, J.; Lenton, P.; Rudney, J.D.; Jones, R.S.; Aparicio, C.; Fok, A. Degradation in the dentin-composite interface subjected to multi-species biofilm challenges. Acta Biomater. 2014, 10, 375–383. [Google Scholar] [CrossRef]
- Pitts, B.; Willse, A.; McFeters, G.A.; Hamilton, M.A.; Zelver, N.; Stewart, P.S. A repeatable laboratory method for testing the efficacy of biocides against toilet bowl biofilms. J. Appl. Microbiol. 2001, 91, 110–117. [Google Scholar] [CrossRef] [Green Version]
- Cotter, J.J.; O’Gara, J.P.; Stewart, P.S.; Pitts, B.; Casey, E. Characterization of a modified rotating disk reactor for the cultivation of Staphylococcus epidermidis biofilm. J. Appl. Microbiol. 2010, 109, 2105–2117. [Google Scholar] [CrossRef]
- Barry, D.M.; McGrath, P.B. Rotation disk process to assess the influence of metals and voltage on the growth of biofilm. Materials 2016, 9, 568. [Google Scholar] [CrossRef]
- Gomes, I.B.; Simoes, M.; Simoes, L.C. An overview on the reactors to study drinking water biofilms. Water Res. 2014, 62, 63–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pintar, K.D.M.; Slawson, R.M. Effect of temperature and disinfection strategies on ammonia-oxidizing bacteria in a bench-scale drinking water distribution system. Water Res. 2003, 37, 1805–1817. [Google Scholar] [CrossRef]
- Ndiongue, S.; Huck, P.M.; Slawson, R.M. Effects of temperature and biodegradable organic matter on control of biofilms by free chlorine in a model drinking water distribution system. Water Res. 2005, 39, 953–964. [Google Scholar] [CrossRef] [PubMed]
- Jang, H.J.; Choi, Y.J.; Ka, J.O. Effects of diverse water pipe materials on bacterial communities and water quality in the annular reactor. J. Microbiol. Biotechnol. 2011, 21, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Goeres, D.M.; Hamilton, M.A.; Beck, N.A.; Buckingham-Meyer, K.; Hilyard, J.D.; Loetterle, L.R.; Lorenz, L.A.; Walker, D.K.; Stewart, P.S. A method for growing a biofilm under low shear at the air-liquid interface using the drip flow biofilm reactor. Nat. Protoc. 2009, 4, 783–788. [Google Scholar] [CrossRef]
- Sawant, S.N.; Selvaraj, V.; Prabhawathi, V.; Doble, M. Antibiofilm properties of silver and gold incorporated PU, PCLm, PC and PMMA nanocomposites under two shear conditions. PLoS ONE 2013, 8, e63311. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, D.G.; Xia, Z.; Gordon, T.B.; Gao, C.; Bouwer, E.J.; Fairbrother, D.H. Biofilm development on carbon nanotube/polymer nanocomposites. Environ. Sci. Nano 2016, 3, 545–558. [Google Scholar] [CrossRef]
- Salli, K.M.; Ouwehand, A.C. The use of in vitro model systems to study dental biofilms associated with caries: A short review. J. Oral Microbiol. 2015, 7, 26149. [Google Scholar] [CrossRef]
- Jaramillo, D.E.; Arriola, A.; Safavi, K.; de Paz, L.E.C. Decreased bacterial adherence and biofilm growth on surfaces coated with a solution of benzalkonium chloride. J. Endod. 2012, 38, 821–825. [Google Scholar] [CrossRef]
- Francolini, I.; Norris, P.; Piozzi, A.; Donelli, G.; Stoodley, P. Usnic acid, a natural antimicrobial agent able to inhibit bacterial biofilm formation on polymer surfaces. Antimicrob. Agents Chemother. 2004, 48, 4360–4365. [Google Scholar] [CrossRef]
- Fabbri, S.; Dennington, S.P.; Price, C.; Stoodley, P.; Longyear, J. A marine biofilm flow cell for in situ screening marine fouling control coatings using optical coherence tomography. Ocean. Eng. 2018, 170, 321–328. [Google Scholar] [CrossRef]
- Tremblay, Y.D.N.; Vogeleer, P.; Jacques, M.; Harel, J. High-throughput microfluidic method to study biofilm formation and host-pathogen interactions in pathogenic Escherichia coli. Appl. Environ. Microbiol. 2015, 81, 2827–2840. [Google Scholar] [CrossRef]
- Benoit, M.R.; Conant, C.G.; Ionescu-Zanetti, C.; Schwartz, M.; Matin, A. New device for high-throughput viability screening of flow biofilms. Appl. Environ. Microbiol. 2010, 76, 4136–4142. [Google Scholar] [CrossRef]
- Moormeier, D.E.; Endres, J.L.; Mann, E.E.; Sadykov, M.R.; Horswill, A.R.; Rice, K.C.; Fey, P.D.; Bayles, K.W. Use of microfluidic technology to analyze gene expression during Staphylococcus aureus biofilm formation reveals distinct physiological niches. Appl. Environ. Microbiol. 2013, 79, 3413–3424. [Google Scholar] [CrossRef]
- Abdulkareem, E.H.; Memarzadeh, K.; Allaker, R.P.; Huang, J.; Pratten, J.; Spratt, D. Anti-biofilm activity of zinc oxide and hydroxyapatite nanoparticles as dental implant coating materials. J. Dent. 2015, 43, 1462–1469. [Google Scholar] [CrossRef]
- Li, F.; Weir, M.D.; Fouad, A.F.; Xu, H.H.K. Effect of salivary pellicle on antibacterial activity of novel antibacterial dental adhesives using a dental plaque microcosm biofilm model. Dent. Mater. 2014, 30, 182–191. [Google Scholar] [CrossRef]
- Wood, T.K.; Knabel, S.J.; Kwan, B.W. Bacterial persister cell formation and dormancy. Appl. Environ. Microbiol. 2013, 79, 7116–7121. [Google Scholar] [CrossRef]
- Hooijmans, C.M.; Abdin, T.A.; Alaerts, G.J. Quantification of viable biomass in biofilm reactors by extractable lipid phosphate. Appl. Microbiol. Biotechnol. 1995, 43, 781–785. [Google Scholar] [CrossRef]
- Quideau, S.A.; McIntosh, A.C.S.; Norris, C.E.; Lloret, E.; Swallow, M.J.B.; Hannam, K. Extraction and analysis of microbial phospholipid fatty acids in soils. J. Vis. Exp. 2016, 114, 54360. [Google Scholar] [CrossRef]
- Willers, C.; van Rensburg, P.J.J.; Claassens, S. Phospholipid fatty acid profiling of microbial communities-a review of interpretations and recent applications. J. Appl. Microbiol. 2015, 119, 1207–1218. [Google Scholar] [CrossRef]
- Gehron, M.J.; White, D.C. Sensitive assay of phospholipid glycerol in environmental sample. J. Microbiol. Met. 1983, 1, 23–32. [Google Scholar] [CrossRef]
- Oursel, D.; Loutelier-Bourhis, C.; Orange, N.; Chevalier, S.; Norris, V.; Lange, C.M. Identification and relative quantification of fatty acids in Escherichia coli membranes by gas chromatography/mass spectrometry. Rapid Commun. Mass Spectrom. 2007, 21, 3229–3233. [Google Scholar] [CrossRef]
- Li, L.; Han, J.J.; Wang, Z.P.; Liu, J.A.; Wei, J.C.; Xiong, S.X.; Zhao, Z.W. Mass spectrometry methodology in lipid analysis. Int. J. Mol. Sci. 2014, 15, 10492–10507. [Google Scholar] [CrossRef]
- Jain, S.; Caforio, A.; Driessen, A.J.M. Biosynthesis of archaeal membrane ether lipids. Front. Microbiol. 2014, 5, 641. [Google Scholar] [CrossRef] [Green Version]
- Gors, S.; Schumann, R.; Haubner, N.; Karsten, U. Fungal and algal biomass in biofilms on artificial surfaces quantified by ergosterol and chlorophyll a as biomarkers. Int. Biodeterior. Biodegrad. 2007, 60, 50–59. [Google Scholar] [CrossRef]
- Hippelein, M.; Rugamer, M. Ergosterol as an indicator of mould growth on building materials. Int. J. Hyg. Environ. Health 2004, 207, 379–385. [Google Scholar] [CrossRef]
- Ng, H.E.; Raj, S.S.A.; Wong, S.H.; Tey, D.; Tan, H.M. Estimation of fungal growth using the ergosterol assay: A rapid tool in assessing the microbiological status of grains and feeds. Lett. Appl. Microbiol. 2008, 46, 113–118. [Google Scholar] [CrossRef]
- Pantanella, F.; Valenti, P.; Natalizi, T.; Passeri, D.; Berlutti, F. Analytical techniques to study microbial biofilm on abiotic surfaces: Pros and cons of the main techniques currently in use. Ann. Ig. 2013, 25, 31–42. [Google Scholar] [CrossRef]
- Welch, K.; Cai, Y.; Strømme, M. A method for quantitative determination of biofilm viability. J. Funct. Biomater. 2012, 3, 418–431. [Google Scholar] [CrossRef]
- Trafny, E.A.; Lewandowski, R.; Zawistowska-Marciniak, I.; Stepinska, M. Use of MTT assay for determination of the biofilm formation capacity of microorganisms in metalworking fluids. World J. Microbiol. Biotechnol. 2013, 29, 1635–1643. [Google Scholar] [CrossRef] [PubMed]
- Nante, N.; Ceriale, E.; Messina, G.; Lenzi, D.; Manzi, P. Effectiveness of ATP bioluminescence to assess hospital cleaning: A review. J. Prev. Med. Hyg. 2017, 58, E177–E183. [Google Scholar] [PubMed]
- AlLuhaybi, K.A.R.; Alghaith, G.Y.; Moneib, N.A.; Yassien, M.A.M. Generation of recombinant bioluminescent Escherichia coli for quantitative determination of bacterial adhesion. Pak. J. Pharm. Sci. 2015, 28, 1301–1306. [Google Scholar] [PubMed]
- Ivanova, E.P.; Alexeeva, Y.V.; Pham, D.K.; Wright, J.P.; Nicolau, D.V. ATP level variations in heterotrophic bacteria during attachment on hydrophilic and hydrophobic surfaces. Int. Microbiol. 2006, 9, 37–46. [Google Scholar] [PubMed]
- Braissant, O.; Wirz, D.; Gopfert, B.; Daniels, A.U. Use of isothermal microcalorimetry to monitor microbial activities. FEMS Microbiol. Lett. 2010, 303, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Solokhina, A.; Bruckner, D.; Bonkat, G.; Braissant, O. Metabolic activity of mature biofilms of Mycobacterium tuberculosis and other non-tuberculous mycobacteria. Sci. Rep. 2017, 7, 9225. [Google Scholar] [CrossRef]
- Said, J.; Walker, M.; Parsons, D.; Stapleton, P.; Beezer, A.E.; Gaisford, S. Development of a flow system for studying biofilm formation on medical devices with microcalorimetry. Methods 2015, 76, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Brueckner, D.; Roesti, D.; Zuber, U.G.; Schmidt, R.; Kraehenbuehl, S.; Bonkat, G.; Braissant, O. Comparison of tunable diode laser absorption spectroscopy and isothermal micro-calorimetry for non-invasive detection of microbial growth in media fills. Sci. Rep. 2016, 6, 27894. [Google Scholar] [CrossRef]
- Wilson, C.; Lukowicz, R.; Merchant, S.; Valquier-Flynn, H.; Caballero, J.; Sandoval, J.; Okuom, M.; Huber, C.; Brooks, T.D.; Wilson, E.; et al. Quantitative and qualitative assessment methods for biofilm growth: A mini-review. Res. Rev. J. Eng. Technol. 2017, 6. [Google Scholar]
- Bogachev, M.I.; Volkov, V.Y.; Markelov, O.A.; Trizna, E.Y.; Baydamshina, D.R.; Melnikov, V.; Murtazina, R.R.; Zelenikhin, P.V.; Sharafutdinov, I.S.; Kayumov, A.R. Fast and simple tool for the quantification of biofilm-embedded cells sub-populations from fluorescent microscopic images. PLoS ONE 2018, 13, e0193267. [Google Scholar] [CrossRef]
- Shi, L.; Gunther, S.; Hubschmann, T.; Wick, L.Y.; Harms, H.; Muller, S. Limits of propidium iodide as a cell viability indicator for environmental bacteria. Cytom. A 2007, 71A, 592–598. [Google Scholar] [CrossRef]
- Netuschil, L.; Auschill, T.M.; Sculean, A.; Arweiler, N.B. Confusion over live/dead stainings for the detection of vital microorganisms in oral biofilms—Which stain is suitable? BMC Oral Health 2014, 14, 2. [Google Scholar] [CrossRef] [PubMed]
- Stiefel, P.; Schmidt-Emrich, S.; Maniura-Weber, K.; Ren, Q. Critical aspects of using bacterial cell viability assays with the fluorophores SYTO9 and propidium iodide. BMC Microbiol. 2015, 15, 36. [Google Scholar] [CrossRef] [PubMed]
- Dobor, J.; Varga, M.; Zaray, G. Biofilm controlled sorption of selected acidic drugs on river sediments characterized by different organic carbon content. Chemosphere 2012, 87, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar]
- Smith, P.K.; Krohn, R.I.; Hermanson, G.T.; Mallia, A.K.; Gartner, F.H.; Provenzano, M.D.; Fujimoto, E.K.; Goeke, N.M.; Olson, B.J.; Klenk, D.C. Measurement of protein using bicinchoninic acid. Anal. Biochem. 1985, 150, 76–85. [Google Scholar] [CrossRef]
- Jesus, B.; Perkins, R.G.; Mendes, C.R.; Brotas, V.; Paterson, D.M. Chlorophyll fluorescence as a proxy for microphytobenthic biomass: Alternatives to the current methodology. Mar. Biol. 2006, 150, 17–28. [Google Scholar] [CrossRef]
- Sanmartin, P.; Aira, N.; Devesa-Rey, R.; Silva, B.; Prieto, B. Relationship between color and pigment production in two stone biofilm-forming cyanobacteria (Nostoc sp PCC 9104 and Nostoc sp PCC 9025). Biofouling 2010, 26, 499–509. [Google Scholar] [CrossRef]
- Sendersky, E.; Simkovsky, R.; Golden, S.S.; Schwarz, R. Quantification of chlorophyll as a proxy for biofilm formation in the cyanobacterium Synechococcus Elongatus. Bio-Protoc. 2017, 7, 14. [Google Scholar] [CrossRef]
- Chamizo, S.; Adessi, A.; Mugnai, G.; Simiani, A.; De Philippis, R. Soil Type and Cyanobacteria species influence the macromolecular and chemical characteristics of the polysaccharidic matrix in induced biocrusts. Microb. Ecol. 2018, 78, 482–493. [Google Scholar] [CrossRef]
- Fernandez-Silva, I.; Sanmartin, P.; Silva, B.; Moldes, A.; Prieto, B. Quantification of phototrophic biomass on rocks: Optimization of chlorophyll-a extraction by response surface methodology. J. Ind. Microbiol. Biotechnol. 2011, 38, 179–188. [Google Scholar] [CrossRef]
- Vazquez-Nion, D.; Silva, B.; Prieto, B. Bioreceptivity index for granitic rocks used as construction material. Sci. Total Environ. 2018, 633, 112–121. [Google Scholar] [CrossRef]
- Vazquez-Nion, D.; Silva, B.; Prieto, B. Influence of the properties of granitic rocks on their bioreceptivity to subaerial phototrophic biofilms. Sci. Total Environ. 2018, 610, 44–54. [Google Scholar] [CrossRef]
- Dalwai, F.; Spratt, D.A.; Pratten, J. Use of quantitative PCR and culture methods to characterize ecological flux in bacterial biofilms. J. Clin. Microbiol. 2007, 45, 3072–3076. [Google Scholar] [CrossRef]
- Alvarez, G.; Gonzalez, M.; Isabal, S.; Blanc, V.; Leon, R. Method to quantify live and dead cells in multi-species oral biofilm by real-time PCR with propidium monoazide. Amb. Express 2013, 3, 1. [Google Scholar] [CrossRef]
- Taylor, M.J.; Bentham, R.H.; Ross, K.E. Limitations of using propidium monoazide with qpcr to discriminate between live and dead Legionella in biofilm samples. Microbiol. Insights 2014, 7, 15–24. [Google Scholar] [CrossRef]
- Soto-Munoz, L.; Teixido, N.; Usall, J.; Vinas, I.; Crespo-Sempere, A.; Torres, R. Development of PMA real-time PCR method to quantify viable cells of Pantoea agglomerans CPA-2, an antagonist to control the major postharvest diseases on oranges. Int. J. Food Microbiol. 2014, 180, 49–55. [Google Scholar] [CrossRef]
- Ambriz-Aviña, V.; Contreras-Garduño, J.A.; Pedraza-Reyes, M. Applications of flow cytometry to characterize bacterial physiological responses. Biomed. Res. Int. 2014, 2014, 461941. [Google Scholar] [CrossRef]
- Kerstens, M.; Boulet, G.; Van Kerckhoven, M.; Clais, S.; Lanckacker, E.; Delputte, P.; Maes, L.; Cos, P. A flow cytometric approach to quantify biofilms. Folia Microbiol. 2015, 60, 335–342. [Google Scholar] [CrossRef]
- Bakke, R.; Kommedal, R.; Kalvenes, S. Quantification of biofilm accumulation by an optical approach. J. Microbiol. Methods 2001, 44, 13–26. [Google Scholar] [CrossRef]
- Stiefel, P.; Rosenberg, U.; Schneider, J.; Mauerhofer, S.; Maniura-Weber, K.; Ren, Q. Is biofilm removal properly assessed? Comparison of different quantification methods in a 96-well plate system. Appl. Microbiol. Biotechnol. 2016, 100, 4135–4145. [Google Scholar] [CrossRef] [Green Version]
- O’Toole, G.A.; Kolter, R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: A genetic analysis. Mol. Microbiol. 1998, 28, 449–461. [Google Scholar] [CrossRef]
- Young, M.E.; Wakefield, R.; Urquhart, D.C.M.; Nicholson, K.; Tonge, K. Assesment in a field setting of the efficacy of various biocides on sandstone. In Int. Coll Methods of Evaluating Products for the Conservation of Porous Building Materials in Monuments; ICCROM: Rome, Italy, 1995; pp. 93–99. ISBN 92-9077-131-3. [Google Scholar]
- Prieto, B.; Rivas, T.; Silva, B. Rapid quantification of phototrophic microorganisms and their physiological state through their colour. Biofouling 2002, 18, 229–236. [Google Scholar] [CrossRef]
- Vazquez-Nion, D.; Sanmartin, P.; Silva, B.; Prieto, B. Reliability of color measurements for monitoring pigment content in a biofilm-forming cyanobacterium. Int. Biodeterior. Biodegrad. 2013, 84, 220–226. [Google Scholar] [CrossRef]
- Sanmartin, P.; Villa, F.; Polo, A.; Silva, B.; Prieto, B.; Cappitelli, F. Rapid evaluation of three biocide treatments against the cyanobacterium Nostoc sp PCC 9104 by color changes. Ann. Microbiol. 2015, 65, 1153–1158. [Google Scholar] [CrossRef]
- Sanmartin, P.; Vazquez-Nion, D.; Arines, J.; Cabo-Dominguez, L.; Prieto, B. Controlling growth and colour of phototrophs by using simple and inexpensive coloured lighting: A preliminary study in the Light4Heritage project towards future strategies for outdoor illumination. Int. Biodeterior. Biodegrad. 2017, 122, 107–115. [Google Scholar] [CrossRef]
- Prieto, B.; Vazquez-Nion, D.; Silva, B.; Sanmartin, P. Shaping colour changes in a biofilm-forming cyanobacterium by modifying the culture conditions. Algal Res. 2018, 33, 173–181. [Google Scholar] [CrossRef]
- Fairchild, M.D. International Commission on Illumination. In CIE 015:2018 Colorimetry, 4th ed.; CIE Central Bureau: Vienna, Austria, 2018; p. 111. ISBN 978-3-902842-13-8. [Google Scholar]
- Sanmartin, P.; Villa, F.; Silva, B.; Cappitelli, F.; Prieto, B. Color measurements as a reliable method for estimating chlorophyll degradation to phaeopigments. Biodegradation 2011, 22, 763–771. [Google Scholar] [CrossRef]
- Di Martino, P. Extracellular polymeric substances, a key element in understanding biofilm phenotype. Aims Microbiol. 2018, 4, 274–288. [Google Scholar] [CrossRef]
- Blanco, Y.; Rivas, L.A.; González-Toril, E.; Ruiz-Bermejo, M.; Moreno-Paz, M.; Parro, V.; Palacín, A.; Aguilera, Á.; Puente-Sánchez, F. Environmental parameters, and not phylogeny, determine the composition of extracellular polymeric substances in microbial mats from extreme environments. Sci. Total Environ. 2019, 650, 384–393. [Google Scholar] [CrossRef]
- Jachlewski, S.; Jachlewski, W.D.; Linne, U.; Bräsen, C.; Wingender, J.; Siebers, B. Isolation of extracellular polymeric substances from biofilms of the thermoacidophilic archaeon Sulfolobus acidocaldarius. Front. Bioeng. Biotechnol. 2015, 3, 123. [Google Scholar] [CrossRef]
- McSwain, B.S.; Irvine, R.L.; Hausner, M.; Wilderer, P.A. Composition and distribution of extracellular polymeric substances in aerobic flocs and granular sludge. Appl. Environ. Microbiol. 2005, 71, 1051–1057. [Google Scholar] [CrossRef]
- Cho, J.; Hermanowicz, S.W.; Hur, J. Effects of experimental conditions on extraction yield of extracellular polymeric substances by cation exchange resin. Sci. World J. 2012, 2012, 751965. [Google Scholar] [CrossRef]
- Rossi, F.; Mugnai, G.; De Philippis, R. Complex role of the polymeric matrix in biological soil crusts. Plant Soil 2018, 429, 19–34. [Google Scholar] [CrossRef]
- Pan, X.L.; Liu, J.; Zhang, D.Y.; Chen, X.; Li, L.H.; Song, W.J.; Yang, J.Y. A comparison of five extraction methods for extracellular polymeric substances (EPS) from biofilm by using three-dimensional excitation-emission matrix (3DEEM) fluorescence spectroscopy. Water Sa 2010, 36, 111–116. [Google Scholar] [CrossRef]
- Liu, H.; Fang, H.H.P. Extraction of extracellular polymeric substances (EPS) of sludges. J. Biotechnol. 2002, 95, 249–256. [Google Scholar] [CrossRef]
- Villa, F.; Remelli, W.; Forlani, F.; Gambino, M.; Landini, P.; Cappitelli, F. Effects of chronic sub-lethal oxidative stress on biofilm formation by Azotobacter vinelandii. Biofouling 2012, 28, 823–833. [Google Scholar] [CrossRef]
- Cattò, C.; Grazioso, G.; Dell’Orto, S.; Gelain, A.; Villa, S.; Marzano, V.; Vitali, A.; Villa, F.; Cappitelli, F.; Forlani, F. The response of Escherichia coli biofilm to salicylic acid. Biofouling 2017, 33, 235–251. [Google Scholar] [CrossRef]
- Masuko, T.; Minami, A.; Iwasaki, N.; Majima, T.; Nishimura, S.I.; Lee, Y.C. Carbohydrate analysis by a phenol-sulfuric acid method in microplate format. Anal. Biochem. 2005, 339, 69–72. [Google Scholar] [CrossRef]
- Nielsen, S.S. Total carbohydrate by phenol-sulfuric acid method. In Food Analysis Laboratory Manual; Springer: Boston, MA, USA, 2010; ISBN 978-1-4419-1462-0. [Google Scholar]
- Ruhmann, B.; Schmid, J.; Sieber, V. Methods to identify the unexplored diversity of microbial exopolysaccharides. Front. Microbiol. 2015, 6, 565. [Google Scholar] [CrossRef] [Green Version]
- Mojica, K.; Elsey, D.; Cooney, M.J. Quantitative analysis of biofilm EPS uronic acid content. J. Microbiol. Methods 2007, 71, 61–65. [Google Scholar] [CrossRef]
- Khodse, V.B.; Bhosle, N.B. Differences in carbohydrate profiles in batch culture grown planktonic and biofilm cells of Amphora rostrata Wm. Sm. Biofouling 2010, 26, 527–537. [Google Scholar] [CrossRef]
- Tielen, P.; Rosenau, F.; Wilhelm, S.; Jaeger, K.E.; Flemming, H.C.; Wingender, J. Extracellular enzymes affect biofilm formation of mucoid Pseudomonas Aeruginosa. Microbiology 2010, 156, 2239–2252. [Google Scholar] [CrossRef]
- Blumenkr, N.; Asboehan, G. New method for quantitative-determination of uronic acids. Anal. Biochem. 1973, 54, 484–489. [Google Scholar] [CrossRef]
- van den Hoogen, B.M.; van Weeren, P.R.; Lopes-Cardozo, M.; van Golde, L.M.G.; Barneveld, A.; van de Lest, C.H.A. A microtiter plate assay for the determination of uronic acids. Anal. Biochem. 1998, 257, 107–111. [Google Scholar] [CrossRef]
- Wu, J.F.; Xi, C.W. Evaluation of different methods for extracting extracellular DNA from the biofilm matrix. Appl. Environ. Microbiol. 2009, 75, 5390–5395. [Google Scholar] [CrossRef]
- Das, T.; Sehar, S.; Manefield, M. The roles of extracellular DNA in the structural integrity of extracellular polymeric substance and bacterial biofilm development. Environ. Microbiol. Rep. 2013, 5, 778–786. [Google Scholar] [CrossRef]
- Steinberger, R.E.; Holden, P.A. Extracellular DNA in single- and multiple-species unsaturated biofilms. Appl. Environ. Microbiol. 2005, 71, 5404–5410. [Google Scholar] [CrossRef]
- Tang, L.; Schramm, A.; Neu, T.R.; Revsbech, N.P.; Meyer, R.L. Extracellular DNA in adhesion and biofilm formation of four environmental isolates: A quantitative study. FEMS Microbiol. Ecol. 2013, 86, 394–403. [Google Scholar] [CrossRef]
- Jiao, Y.Q.; Cody, G.D.; Harding, A.K.; Wilmes, P.; Schrenk, M.; Wheeler, K.E.; Banfield, J.F.; Thelen, M.P. Characterization of extracellular polymeric substances from acidophilic microbial biofilms. Appl. Environ. Microbiol. 2010, 76, 2916–2922. [Google Scholar] [CrossRef]
- Nan, L.; Yang, K.; Ren, G.G. Anti-biofilm formation of a novel stainless steel against Staphylococcus aureus. Mat. Sci. Eng. C 2015, 51, 356–361. [Google Scholar] [CrossRef]
- Kelestemur, S.; Avci, E.; Culha, M. Raman and surface-enhanced raman scattering for biofilm characterization. Chemosensors 2018, 6, 5. [Google Scholar] [CrossRef]
- Ramirez-Mora, T.; Davila-Perez, C.; Torres-Mendez, F.; Valle-Bourrouet, G. Raman spectroscopic characterization of endodontic biofilm matrices. J. Spectrosc. 2019, 2019, 1307397. [Google Scholar] [CrossRef]
- Chao, Y.Q.; Zhang, T. Surface-enhanced Raman scattering (SERS) revealing chemical variation during biofilm formation: From initial attachment to mature biofilm. Anal. Bioanal. Chem. 2012, 404, 1465–1475. [Google Scholar] [CrossRef]
- Xu, H.C.; He, P.J.; Wang, G.Z.; Shao, L.M. Three-dimensional excitation emission matrix fluorescence spectroscopy and gel-permeating chromatography to characterize extracellular polymeric substances in aerobic granulation. Water Sci. Technol. 2010, 61, 2931–2942. [Google Scholar] [CrossRef]
- Bales, P.M.; Renke, E.M.; May, S.L.; Shen, Y.; Nelson, D.C. Purification and characterization of biofilm-associated eps exopolysaccharides from ESKAPE organisms and other pathogens. PLoS ONE 2013, 8, e67950. [Google Scholar] [CrossRef]
- Ruhmann, B.; Schmid, J.; Sieber, V. Fast carbohydrate analysis via liquid chromatography coupled with ultra violet and electrospray ionization ion trap detection in 96-well format. J. Chromatogr. A 2014, 1350, 44–50. [Google Scholar] [CrossRef]
- Ramirez-Mora, T.; Retana-Lobo, C.; Valle-Bourrouet, G. Biochemical characterization of extracellular polymeric substances from endodontic biofilms. PLoS ONE 2018, 13, e0204081. [Google Scholar] [CrossRef]
- Hasan, N.; Gopal, J.; Wu, H.F. Rapid, sensitive and direct analysis of exopolysaccharides from biofilm on aluminum surfaces exposed to sea water using MALDI-TOF MS. J. Mass Spectrom. 2011, 46, 1160–1167. [Google Scholar] [CrossRef]
- Gonzalez-Gil, G.; Thomas, L.; Emwas, A.H.; Lens, P.N.; Saikaly, P.E. NMR and MALDI-TOF MS based characterization of exopolysaccharides in anaerobic microbial aggregates from full-scale reactors. Sci. Rep. 2015, 5, 14316. [Google Scholar] [CrossRef]
- Yildiz, F.; Fong, J.; Sadovskaya, I.; Grard, T.; Vinogradov, E. Structural characterization of the extracellular polysaccharide from Vibrio cholerae O1 El-Tor. PLoS ONE 2014, 9, e86751. [Google Scholar] [CrossRef]
- Neu, T.R.; Lawrence, J.R. Advanced techniques for in situ analysis of the biofilm matrix (structure, composition, dynamics) by means of laser scanning microscopy. Methods Mol. Biol. 2014, 1147, 43–64. [Google Scholar] [CrossRef]
- Cowan, S.E.; Gilbert, E.; Liepmann, D.; Keasling, J.D. Commensal interactions in a dual-species biofilm exposed to mixed organic compounds. Appl. Environ. Microbiol. 2000, 66, 4481–4485. [Google Scholar] [CrossRef]
- Chen, M.Y.; Lee, D.J.; Tay, J.H.; Show, K.Y. Staining of extracellular polymeric substances and cells in bioaggregates. Appl. Microbiol. Biotechnol. 2007, 75, 467–474. [Google Scholar] [CrossRef]
- Berk, V.; Fong, J.C.N.; Dempsey, G.T.; Develioglu, O.N.; Zhuang, X.W.; Liphardt, J.; Yildiz, F.H.; Chu, S. Molecular architecture and assembly principles of Vibrio cholerae biofilms. Science 2012, 337, 236–239. [Google Scholar] [CrossRef]
- Sportelli, M.C.; Tutuncu, E.; Picca, R.A.; Valentini, M.; Valentini, A.; Kranz, C.; Mizaikoff, B.; Barth, H.; Cioffi, N. Inhibiting, P. Fluorescens biofilms with fluoropolymer-embedded silver nanoparticles: An in-situ spectroscopic study. Sci. Rep. 2017, 7, 11870. [Google Scholar] [CrossRef]
- Feng, J.; de la Fuente-Núñez, C.; Trimble, M.J.; Xu, J.; Hancock, R.E.; Lu, X. An in situ Raman spectroscopy-based microfluidic “lab-on-a-chip” platform for non-destructive and continuous characterization of Pseudomonas aeruginosa biofilms. Chem. Commun. 2015, 51, 8966–8969. [Google Scholar] [CrossRef]
- Greuter, D.; Loy, A.; Horne, M.; Ratteil, T. probeBase-an online resource for rRNA-targeted oligonucleotide probes and primers: New features 2016. Nucleic Acids Res. 2016, 44, D586–D589. [Google Scholar] [CrossRef]
- Schimak, M.P.; Kleiner, M.; Wetzel, S.; Liebeke, M.; Dubilier, N.; Fuchs, B.M. MiL-FISH: Multilabeled oligonucleotides for fluorescence in situ hybridization improve visualization of bacterial cells. Appl. Environ. Microbiol. 2016, 82, 62–70. [Google Scholar] [CrossRef]
- Almeida, C.; Azevedo, N.F.; Santos, S.; Keevil, C.W.; Vieira, M.J. Discriminating multi-species populations in biofilms with peptide nucleic acid fluorescence in situ hybridization (PNA FISH). PLoS ONE 2011, 6, e14786. [Google Scholar] [CrossRef]
- Azevedo, A.S.; Almeida, C.; Pereira, B.; Madureira, P.; Wengel, J.; Azevedo, N.F. Detection and discrimination of biofilm populations using locked nucleic acid/2 ‘-O-methyl-RNA fluorescence in situ hybridization (LNA/2 ‘ OMe-FISH). Biochem. Eng. J. 2015, 104, 64–73. [Google Scholar] [CrossRef]
- Kubota, K. CARD-FISH for environmental microorganisms: Technical advancement and future applications. Microbes Environ. 2013, 28, 3–12. [Google Scholar] [CrossRef]
- Escudero, C.; Vera, M.; Oggerin, M.; Amils, R. Active microbial biofilms in deep poor porous continental subsurface rocks. Sci. Rep. 2018, 8, 1538. [Google Scholar] [CrossRef]
- Stoecker, K.; Dorninger, C.; Daims, H.; Wagner, M. Double labeling of oligonucleotide probes for fluorescence in situ hybridization (DOPE-FISH) improves signal intensity and increases rRNA accessibility. Appl. Environ. Microbiol. 2010, 76, 922–926. [Google Scholar] [CrossRef]
- Valm, A.M.; Oldenbourg, R.; Borisy, G.G. Multiplexed spectral imaging of 120 different fluorescent labels. PLoS ONE 2016, 11, e0158495. [Google Scholar] [CrossRef]
- Valm, A.M.; Welch, J.L.M.; Borisy, G.G. CLASI-FISH: Principles of combinatorial labeling and spectral imaging. Syst. Appl. Microbiol. 2012, 35, 496–502. [Google Scholar] [CrossRef] [Green Version]
- Congestri, R. FISH methods in phycology: Phototrophic biofilm and phytoplankton applications. Plant Biosyst. 2008, 142, 337–342. [Google Scholar] [CrossRef]
- Musat, N.; Foster, R.; Vagner, T.; Adam, B.; Kuypers, M.M.M. Detecting metabolic activities in single cells, with emphasis on nanoSIMS. FEMS Microbiol. Rev. 2012, 36, 486–511. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Wu, T.D.; Mazeas, L.; Toffin, L.; Guerquin-Kern, J.L.; Leblon, G.; Bouchez, T. Simultaneous analysis of microbial identity and function using NanoSIMS. Environ. Microbiol. 2008, 10, 580–588. [Google Scholar] [CrossRef] [Green Version]
- Kniggendorf, A.K.; Nogueira, R.; Kelb, C.; Schadzek, P.; Meinhardt-Wollweber, M.; Ngezahayo, A.; Roth, B. Confocal Raman microscopy and fluorescent in situ hybridization-a complementary approach for biofilm analysis. Chemosphere 2016, 161, 112–118. [Google Scholar] [CrossRef]
- Manti, A.; Boi, P.; Amalfitano, S.; Puddu, A.; Papa, S. Experimental improvements in combining CARD-FISH and flow cytometry for bacterial cell quantification. J. Microbiol. Methods 2011, 87, 309–315. [Google Scholar] [CrossRef]
- Janse, J.D.; Kokoskova, B. Indirect immunofluorescence microscopy for the detection and identification of plant pathogenic bacteria (in particular for Ralstonla solanacearum). Methods Mol. Biol. 2009, 508, 89–99. [Google Scholar]
- Bruneval, P.; Choucair, J.; Paraf, F.; Casalta, J.P.; Raoult, D.; Scherchen, F.; Mainardi, J.L. Detection of fastidious bacteria in cardiac valves in cases of blood culture negative endocarditis. J. Clin. Pathol. 2001, 54, 238–240. [Google Scholar] [CrossRef] [Green Version]
- Lin, M.; Todoric, D.; Mallory, M.; Luo, B.S.; Trottier, E.; Dan, H.H. Monoclonal antibodies binding to the cell surface of Listeria monocytogenes serotype 4b. J. Med. Microbiol. 2006, 55, 291–299. [Google Scholar] [CrossRef]
- Foulston, L.; Elsholz, A.K.W.; DeFrancesco, A.S.; Losick, R. The extracellular matrix of Staphylococcus aureus biofilms comprises cytoplasmic proteins that associate with the cell surface in response to decreasing pH. MBio 2014, 5, e01667-14. [Google Scholar] [CrossRef]
- Vejborg, R.M.; Klemm, P. Cellular chain formation in Escherichia coli biofilms. Microbiology 2009, 155, 1407–1417. [Google Scholar] [CrossRef]
- Luo, T.L.; Eisenberg, M.C.; Hayashi, M.A.L.; Gonzalez-Cabezas, C.; Foxman, B.; Marrs, C.F.; Rickard, A.H. A Sensitive thresholding method for confocal laser scanning microscope image stacks of microbial biofilms. Sci. Rep. 2018, 8, 13013. [Google Scholar] [CrossRef]
- Heydorn, A.; Nielsen, A.T.; Hentzer, M.; Sternberg, C.; Givskov, M.; Ersboll, B.K.; Molin, S. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 2000, 146, 2395–2407. [Google Scholar] [CrossRef]
- Mueller, L.N.; de Brouwer, J.F.; Almeida, J.S.; Stal, L.J.; Xavier, J.B. Analysis of a marine phototrophic biofilm by confocal laser scanning microscopy using the new image quantification software PHLIP. BMC Ecol. 2006, 6, 1. [Google Scholar] [CrossRef]
- Sommerfeld Ross, S.S.; Tu, M.H.; Falsetta, M.L.; Ketterer, M.R.; Kiedrowski, M.R.; Horswill, A.R.; Apicella, M.A.; Reinhardt, J.M.; Fiegel, J. Quantification of confocal images of biofilms grown on irregular surfaces. J. Microbiol. Methods 2014, 100, 111–120. [Google Scholar] [CrossRef] [Green Version]
- Miura, K. Bioimage Data Analysis, 1st ed.; Wiley-VCH Verlag GmbH & Co KGaA: Weinheim, Germany, 2016; ISBN 978-3-527-80092-6. [Google Scholar]
- Neu, T.R.; Manz, B.; Volke, F.; Dynes, J.J.; Hitchcock, A.P.; Lawrence, J.R. Advanced imaging techniques for assessment of structure, composition and function in biofilm systems. FEMS Microbiol. Ecol. 2010, 72, 1–21. [Google Scholar] [CrossRef]
- Oubekka, S.D.; Briandet, R.; Fontaine-Aupart, M.P.; Steenkeste, K. Correlative time-resolved fluorescence microscopy to assess antibiotic diffusion-reaction in biofilms. Antimicrob. Agents Chemother. 2012, 56, 3349–3358. [Google Scholar] [CrossRef]
- Davison, W.M.; Pitts, B.; Stewart, P.S. Spatial and temporal patterns of biocide action against Staphylococcus epidermidis biofilms. Antimicrob. Agents Chemother. 2010, 54, 2920–2927. [Google Scholar] [CrossRef]
- Cattò, C.; James, G.; Villa, F.; Villa, S.; Cappitelli, F. Zosteric acid and salicylic acid bound to a low density polyethylene surface successfully control bacterial biofilm formation. Biofouling 2018, 34, 440–452. [Google Scholar] [CrossRef]
- Schneider, J.P.; Basler, M. Shedding light on biology of bacterial cells. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2016, 371, 20150499. [Google Scholar] [CrossRef] [Green Version]
- Power, R.M.; Huisken, J. A guide to light-sheet fluorescence microscopy for multiscale imaging. Nat. Methods 2017, 14, 360–373. [Google Scholar] [CrossRef]
- Parthasarathy, R. Monitoring microbial communities using light sheet fluorescence microscopy. Curr. Opin. Microbiol. 2018, 43, 31–37. [Google Scholar] [CrossRef]
- Janissen, R.; Murillo, D.M.; Niza, B.; Sahoo, P.K.; Nobrega, M.M.; Cesar, C.L.; Temperini, M.L.A.; Carvalho, H.F.; de Souza, A.A.; Cotta, M.A. Spatiotemporal distribution of different extracellular polymeric substances and filamentation mediate Xylella fastidiosa adhesion and biofilm formation. Sci. Rep. 2015, 5, 9856. [Google Scholar] [CrossRef] [Green Version]
- Yan, J.; Sharo, A.G.; Stone, H.A.; Wingreen, N.S.; Bassler, B.L. Vibrio cholerae biofilm growth program and architecture revealed by single-cell live imaging. Proc. Natl. Acad. Sci. USA 2016, 113, E5337–E5343. [Google Scholar] [CrossRef]
- Bryers, J.D. Two-photon excitation microscopy for analyses of biofilm processes. Methods Enzymol. 2001, 337, 259–269. [Google Scholar] [CrossRef]
- Thomsen, H.; Graf, F.E.; Farewell, A.; Ericson, M.B. Exploring photoinactivation of microbial biofilms using laser scanning microscopy and confined 2-photon excitation. J. Biophotonics 2018, 11, e201800018. [Google Scholar] [CrossRef] [Green Version]
- Villa, F.; Giacomucci, L.; Polo, A.; Principi, P.; Toniolo, L.; Levi, M.; Turri, S.; Cappitelli, F. N-vanillylnonanamide tested as a non-toxic antifoulant, applied to surfaces in a polyurethane coating. Biotechnol. Lett. 2009, 31, 1407–1413. [Google Scholar] [CrossRef]
- Trentin, D.S.; Silva, D.B.; Frasson, A.P.; Rzhepishevska, O.; da Silva, M.V.; Pulcini, E.E.L.; James, G.; Soares, G.V.; Tasca, T.; Ramstedt, M.; et al. Natural Green coating inhibits adhesion of clinically important bacteria. Sci. Rep. 2015, 5, 8287. [Google Scholar] [CrossRef]
- Das Ghatak, P.; Mathew-Steiner, S.S.; Pandey, P.; Roy, S.; Sen, C.K. A surfactant polymer dressing potentiates antimicrobial efficacy in biofilm disruption. Sci. Rep. 2018, 8, 873. [Google Scholar] [CrossRef]
- Akuzov, D.; Franca, L.; Grunwald, I.; Vladkova, T. Sharply reduced biofilm formation from cobetia marina and in black sea water on modified siloxane coatings. Coatings 2018, 8, 136. [Google Scholar] [CrossRef]
- Ivanova, E.P.; Hasan, J.; Webb, H.K.; Gervinskas, G.; Juodkazis, S.; Truong, V.K.; Wu, A.H.F.; Lamb, R.N.; Baulin, V.A.; Watson, G.S.; et al. Bactericidal activity of black silicon. Nat. Commun. 2013, 4, 2838. [Google Scholar] [CrossRef]
- Li, L.; Molin, S.; Yang, L.; Ndoni, S. Sodium dodecyl sulfate (SDS)-loaded nanoporous polymer as anti-biofilm surface coating material. Int. J. Mol. Sci. 2013, 14, 3050–3064. [Google Scholar] [CrossRef]
- Dickson, M.N.; Liang, E.I.; Rodriguez, L.A.; Vollereaux, N.; Yee, A.F. Nanopatterned polymer surfaces with bactericidal properties. Biointerphases 2015, 10, 021010. [Google Scholar] [CrossRef] [Green Version]
- Ho, K.K.K.; Ozcelik, B.; Willcox, M.D.P.; Thissen, H.; Kumar, N. Facile solvent-free fabrication of nitric oxide (NO)-releasing coatings for prevention of biofilm formation. Chem. Commun. 2017, 53, 6488–6491. [Google Scholar] [CrossRef]
- Xu, Y.C.; Wang, J.Z.; Hao, Z.Y.; Wang, S.; Liang, C.Z. Biodegradable ciprofloxacin-incorporated waterborne polyurethane polymers prevent bacterial biofilm formation in vitro. Exp. Ther. Med. 2019, 17, 1831–1836. [Google Scholar] [CrossRef]
- Valencia, L.; Kumar, S.; Jalvo, B.; Mautner, A.; Salazar-Alvarez, G.; Mathew, A.P. Fully bio-based zwitterionic membranes with superior antifouling and antibacterial properties prepared via surface-initiated free-radical polymerization of poly(cysteine methacrylate). J. Mater. Chem. A 2018, 6, 16277–16712. [Google Scholar] [CrossRef]
- Dave, R.N.; Joshi, H.M.; Venugopalan, V.P. Novel biocatalytic polymer-based antimicrobial coatings as potential ureteral biomaterial: Preparation and in vitro performance evaluation. Antimicrob. Agents Chemother. 2011, 55, 845–853. [Google Scholar] [CrossRef]
- Sabatini, V.; Cattò, C.; Cappelletti, G.; Cappitelli, F.; Antenucci, S.; Farina, H.; Ortenzi, M.A.; Camazzola, S.; Di Silvestro, G. Protective features, durability and biodegration study of acrylic and methacrylic fluorinated polymer coatings for marble protection. Prog. Org. Coat. 2018, 114, 47–57. [Google Scholar] [CrossRef]
- Albright, V.; Zhuk, I.; Wang, Y.H.; Selin, V.; van de Belt-Gritter, B.; Busscher, H.J.; van der Mei, H.C.; Sukhishvili, S.A. Self-defensive antibiotic-loaded layer-by-layer coatings: Imaging of localized bacterial acidification and pH-triggering of antibiotic release. Acta Biomater. 2017, 61, 66–74. [Google Scholar] [CrossRef]
- Lagree, K.; Mon, H.H.; Mitchell, A.P.; Ducker, W.A. Impact of surface topography on biofilm formation by Candida albicans. PLoS ONE 2018, 13, e0197925. [Google Scholar] [CrossRef]
- Alhede, M.; Qvortrup, K.; Liebrechts, R.; Hoiby, N.; Givskov, M.; Bjarnsholt, T. Combination of microscopic techniques reveals a comprehensive visual impression of biofilm structure and composition. FEMS Immunol. Med. Microbiol. 2012, 65, 335–342. [Google Scholar] [CrossRef] [Green Version]
- Sugimoto, S.; Okuda, K.; Miyakawa, R.; Sato, M.; Arita-Morioka, K.; Chiba, A.; Yamanaka, K.; Ogura, T.; Mizunoe, Y.; Sato, C. Imaging of bacterial multicellular behaviour in biofilms in liquid by atmospheric scanning electron microscopy. Sci. Rep. 2016, 6, 25889. [Google Scholar] [CrossRef] [Green Version]
- Bridier, A.; Meylheuc, T.; Briandet, R. Realistic representation of Bacillus subtilis biofilms architecture using combined microscopy (CLSM, ESEM and FESEM). Micron 2013, 48, 65–69. [Google Scholar] [CrossRef]
- Asahi, Y.; Miura, J.; Tsuda, T.; Kuwabata, S.; Tsunashima, K.; Noiri, Y.; Sakata, T.; Ebisu, S.; Hayashi, M. Simple observation of Streptococcus mutans biofilm by scanning electron microscopy using ionic liquids. AMB Express 2015, 5, 6. [Google Scholar] [CrossRef]
- Gonzalez-Ramirez, A.I.; Ramirez-Granillo, A.; Medina-Canales, M.G.; Rodriguez-Tovar, A.V.; Martinez-Rivera, M.A. Analysis and description of the stages of Aspergillus fumigatus biofilm formation using scanning electron microscopy. BMC Microbiol. 2016, 16, 243. [Google Scholar] [CrossRef]
- Gomes, L.C.; Mergulhao, F.J. SEM analysis of surface impact on biofilm antibiotic treatment. Scanning 2017, 2017, 2960194. [Google Scholar] [CrossRef]
- Mohmmed, S.A.; Vianna, M.E.; Penny, M.R.; Hilton, S.T.; Mordan, N.; Knowles, J.C. Confocal laser scanning, scanning electron, and transmission electron microscopy investigation of Enterococcus faecalis biofilm degradation using passive and active sodium hypochlorite irrigation within a simulated root canal model. Microbiologyopen 2017, 6, e00455. [Google Scholar] [CrossRef]
- McCutcheon, J.; Southam, G. Advanced biofilm staining techniques for TEM and SEM in geomicrobiology: Implications for visualizing EPS architecture, mineral nucleation, and microfossil generation. Chem. Geol. 2018, 498, 115–127. [Google Scholar] [CrossRef]
- Hrubanova, K.; Nebesarova, J.; Ruzicka, F.; Krzyzanek, V. The innovation of cryo-SEM freeze-fracturing methodology demonstrated on high pressure frozen biofilm. Micron 2018, 110, 28–35. [Google Scholar] [CrossRef]
- Liu, M.H.; Wu, X.X.; Li, J.K.; Liu, L.; Zhang, R.G.; Shao, D.Y.; Du, X.D. The specific anti-biofilm effect of gallic acid on Staphylococcus aureus by regulating the expression of the ica operon. Food Control 2017, 73, 613–618. [Google Scholar] [CrossRef]
- Nishiyama, H.; Koizumi, M.; Ogawa, K.; Kitamura, S.; Konyuba, Y.; Watanabe, Y.; Ohbayashi, N.; Fukuda, M.; Suga, M.; Sato, C. Atmospheric scanning electron microscope system with an open sample chamber: Configuration and applications. Ultramicroscopy 2014, 147, 86–97. [Google Scholar] [CrossRef]
- Sahl, S.J.; Hell, S.W.; Jakobs, S. Fluorescence nanoscopy in cell biology. Nat. Rev. Mol. Cell. Biol. 2017, 18, 685–701. [Google Scholar] [CrossRef]
- Blom, H.; Widengren, J. Stimulated emission depletion microscopy. Chem. Rev. 2017, 117, 7377–7427. [Google Scholar] [CrossRef]
- Coltharp, C.; Xiao, J. Superresolution microscopy for microbiology. Cell Microbiol. 2012, 14, 1808–1818. [Google Scholar] [CrossRef] [Green Version]
- Xiao, J.; Dufrêne, Y.F. Optical and force nanoscopy in microbiology. Nat. Microbiol. 2016, 1, 16186. [Google Scholar] [CrossRef]
- Li, C.K.; Kuang, C.F.; Liu, X. Prospects for fluorescence nanoscopy. ACS Nano 2018, 12, 4081–4085. [Google Scholar] [CrossRef]
- Sydor, A.M.; Czymmek, K.J.; Puchner, E.M.; Mennella, V. Super-resolution microscopy: From single molecules to supramolecular assemblies. Trends Cell. Biol. 2015, 25, 730–748. [Google Scholar] [CrossRef]
- Ball, G.; Demmerle, J.; Kaufmann, R.; Davis, I.; Dobbie, I.M.; Schermelleh, L. SIMcheck: A toolbox for successful super-resolution structured illumination microscopy. Sci. Rep. 2015, 5, 15915. [Google Scholar] [CrossRef]
- Virdis, B.; Harnisch, F.; Batstone, D.J.; Rabaey, K.; Donose, B.C. Non-invasive characterization of electrochemically active microbial biofilms using confocal Raman microscopy. Energy Environ. Sci. 2012, 5, 7017–7024. [Google Scholar] [CrossRef]
- Wagner, M.; Horn, H. Optical coherence tomography in biofilm research: A comprehensive review. Biotechnol. Bioeng. 2017, 114, 1386–1402. [Google Scholar] [CrossRef]
- Sandt, C.; Smith-Palmer, T.; Comeau, J.; Pink, D. Quantification of water and biomass in small colony variant PAO1 biofilms by confocal Raman microspectroscopy. Appl. Microbiol. Biotechnol. 2009, 83, 1171–1182. [Google Scholar] [CrossRef]
- Sandt, C.; Smith-Palmer, T.; Pink, J.; Brennan, L.; Pink, D. Confocal Raman microspectroscopy as a tool for studying the chemical heterogeneities of biofilms in situ. J. Appl. Microbiol. 2007, 103, 1808–1820. [Google Scholar] [CrossRef]
- Wagner, M.; Ivleva, N.P.; Haisch, C.; Niessner, R.; Horn, H. Combined use of confocal laser scanning microscopy (CLSM) and Raman microscopy (RM): Investigations on EPS-Matrix. Water Res. 2009, 43, 63–76. [Google Scholar] [CrossRef]
- Andrews, J.S.; Rolfe, S.A.; Huang, W.E.; Scholes, J.D.; Banwart, S.A. Biofilm formation in environmental bacteria is influenced by different macromolecules depending on genus and species. Environ. Microbiol. 2010, 12, 2496–2507. [Google Scholar] [CrossRef]
- Lawrence, J.R.; Swerhone, G.D.W.; Dynes, J.J.; Hitchcock, A.P.; Korber, D.R. Complex organic corona formation on carbon nanotubes reduces microbial toxicity by suppressing reactive oxygen species production. Environ. Sci. Nano 2016, 3, 181–189. [Google Scholar] [CrossRef]
- Yang, S.I.; George, G.N.; Lawrence, J.R.; Kaminskyj, S.G.W.; Dynes, J.J.; Lai, B.; Pickering, I.J. Multispecies biofilms transform selenium oxyanions into elemental selenium particles: Studies using combined synchrotron x-ray fluorescence imaging and scanning transmission x-ray microscopy. Environ. Sci. Technol. 2016, 50, 10343–10350. [Google Scholar] [CrossRef]
- Blauert, F.; Horn, H.; Wagner, M. Time-resolved biofilm deformation measurements using optical coherence tomography. Biotechnol. Bioeng. 2015, 112, 1893–1905. [Google Scholar] [CrossRef]
- Heidari, A.E.; Moghaddam, S.; Truong, K.K.; Chou, L.; Genberg, C.; Brenner, M.; Chena, Z.P. Visualizing biofilm formation in endotracheal tubes using endoscopic three-dimensional optical coherence tomography. J. Biomed. Opt. 2016, 21, 126010. [Google Scholar] [CrossRef]
- Farid, M.U.; Guo, J.X.; An, A.K. Bacterial inactivation and in situ monitoring of biofilm development on graphene oxide membrane using optical coherence tomography. J. Memb. Sci. 2018, 564, 22–34. [Google Scholar] [CrossRef]
- Dreszer, C.; Wexler, A.D.; Drusova, S.; Overdijk, T.; Zwijnenburg, A.; Flemming, H.C.; Kruithof, J.C.; Vrouwenvelder, J.S. In-situ biofilm characterization in membrane systems using optical coherence tomography: Formation, structure, detachment and impact of flux change. Water Res. 2014, 67, 243–254. [Google Scholar] [CrossRef]
- Fortunato, L.; Jeong, S.; Leiknes, T. Time-resolved monitoring of biofouling development on a flat sheet membrane using optical coherence tomography. Sci. Rep. 2017, 7, 15. [Google Scholar] [CrossRef]
- Ogrodzki, P.; Cheung, C.S.; Saad, M.; Dahmani, K.; Coxill, R.; Liang, H.D.; Forsythe, S.J. Rapid in situ imaging and whole genome sequencing of biofilm in neonatal feeding tubes: A clinical proof of concept. Sci. Rep. 2017, 7, 15948. [Google Scholar] [CrossRef] [Green Version]
- de Andrade, M.C.L.; de Oliveira, M.A.S.; dos Santos, F.D.G.; Vilela, P.D.X.; da Silva, M.N.; Macedo, D.P.C.; Neto, R.G.D.; Neves, H.J.P.; Brandao, I.D.L.; Chaves, G.M.; et al. A new approach by optical coherence tomography for elucidating biofilm formation by emergent Candida species. PLoS ONE 2017, 12, e0188020. [Google Scholar] [CrossRef]
- Rupp, C.J.; Fux, C.A.; Stoodley, P. Viscoelasticity of Staphylococcus aureus biofilms in response to fluid shear allows resistance to detachment and facilitates rolling migration. Appl. Environ. Microbiol. 2005, 71, 2175–2178. [Google Scholar] [CrossRef]
- Kim, M.K.; Drescher, K.; Pak, O.S.; Bassler, B.L.; Stone, H.A. Filaments in curved streamlines: Rapid formation of Staphylococcus aureus biofilm streamers. New J. Phys. 2014, 16, 065024. [Google Scholar] [CrossRef]
- Billings, N.; Birjiniuk, A.; Samad, T.S.; Doyle, P.S.; Ribbeck, K. Material properties of biofilms—A review of methods for understanding permeability and mechanics. Rep. Progr. Phys. 2015, 78, 036601. [Google Scholar] [CrossRef]
- Boudarel, H.; Mathias, J.D.; Blaysat, B.; Grediac, M. Towards standardized mechanical characterization of microbial biofilms: Analysis and critical review. NPJ Biofilms Microbiomes 2018, 4, 17. [Google Scholar] [CrossRef]
- Bol, M.; Ehret, A.E.; Albero, A.B.; Hellriegel, J.; Krull, R. Recent advances in mechanical characterisation of biofilm and their significance for material modelling. Crit. Rev. Biotechnol. 2013, 33, 145–171. [Google Scholar] [CrossRef]
- Martin, K.J.; Bolster, D.; Derlon, N.; Morgenroth, E.; Nerenberg, R. Effect of fouling layer spatial distribution on permeate flux: A theoretical and experimental study. J. Membr. Sci. 2014, 471, 130–137. [Google Scholar] [CrossRef]
- Li, C.Y.; Wagner, M.; Lackner, S.; Horn, H. Assessing the influence of biofilm surface roughness on mass transfer by combining optical coherence tomography and two-dimensional modeling. Biotechnol. Bioeng. 2016, 113, 989–1000. [Google Scholar] [CrossRef]
- Jafari, M.; Desmond, P.; van Loosdrecht, M.C.M.; Derlon, N.; Morgenroth, E.; Picioreanu, C. Effect of biofilm structural deformation on hydraulic resistance during ultrafiltration: A numerical and experimental study. Water Res. 2018, 145, 375–387. [Google Scholar] [CrossRef]
- Picioreanu, C.; Blauert, F.; Horn, H.; Wagner, M. Determination of mechanical properties of biofilms by modelling the deformation measured using optical coherence tomography. Water Res. 2018, 145, 588–598. [Google Scholar] [CrossRef]
- Dufrene, Y.F. Sticky microbes: Forces in microbial cell adhesion. Trends Microbiol. 2015, 23, 376–382. [Google Scholar] [CrossRef]
- James, S.A.; Powell, L.C.; Wright, C.J. Atomic force microscopy of biofilms-imaging, interactions, and mechanics. In Microbial Biofilms—Importance and Applications; Dhanasekaran, D., Thajuddin, N., Eds.; IntechOpen limited: London, UK, 2016; pp. 95–118. [Google Scholar] [CrossRef]
- Lau, P.C.; Dutcher, J.R.; Beveridge, T.J.; Lam, J.S. Absolute quantitation of bacterial biofilm adhesion and viscoelasticity by microbead force spectroscopy. Biophys. J. 2009, 96, 2935–2948. [Google Scholar] [CrossRef]
- Harapanahalli, A.K.; Chen, Y.; Li, J.; Busscher, H.J.; van der Mei, H.C. Influence of adhesion force on icaA and cidA Gene expression and production of matrix components in Staphylococcus aureus biofilms. Appl. Environ. Microbiol. 2015, 81, 3369–3378. [Google Scholar] [CrossRef]
- Feuillie, C.; Formosa-Dague, C.; Hays, L.M.; Vervaeck, O.; Derclaye, S.; Brennan, M.P.; Foster, T.J.; Geoghegan, J.A.; Dufrêne, Y.F. Molecular interactions and inhibition of the staphylococcal biofilm-forming protein SdrC. Proc. Natl. Acad. Sci. USA 2017, 114, 3738–3743. [Google Scholar] [CrossRef] [Green Version]
- El-Kirat-Chatel, S.; Puymege, A.; Duong, T.H.; Van Overtvelt, P.; Bressy, C.; Belec, L.; Dufrêne, Y.F.; Molmeret, M. Phenotypic heterogeneity in attachment of marine bacteria toward antifouling copolymers unraveled by AFM. Front. Microbiol. 2017, 8, 1399. [Google Scholar] [CrossRef]
- Kundukad, B.; Seviour, T.; Liang, Y.; Rice, S.A.; Kjelleberg, S.; Doyle, P.S. Mechanical properties of the superficial biofilm layer determine the architecture of biofilms. Soft Matter 2016, 12, 5718–5726. [Google Scholar] [CrossRef] [Green Version]
- Taubenberger, A.V.; Hutmacher, D.W.; Muller, D.J. Single-cell force spectroscopy, an emerging tool to quantify cell adhesion to biomaterials. Tissue Eng. Part B Rev. 2014, 20, 40–55. [Google Scholar] [CrossRef]
- Spengler, C.; Thewes, N.; Jung, P.; Bischoff, M.; Jacobs, K. Determination of the nano-scaled contact area of staphylococcal cells. Nanoscale 2017, 9, 10084–10093. [Google Scholar] [CrossRef]
- Klapper, I.; Rupp, C.J.; Cargo, R.; Purvedorj, B.; Stoodley, P. Viscoelastic fluid description of bacterial biofilm material properties. Biotechnol. Bioeng. 2002, 80, 289–296. [Google Scholar] [CrossRef]
- Peterson, B.W.; He, Y.; Ren, Y.; Zerdoum, A.; Libera, M.R.; Sharma, P.K.; van Winkelhoff, A.J.; Neut, D.; Stoodley, P.; van der Mei, H.C.; et al. Viscoelasticity of biofilms and their recalcitrance to mechanical and chemical challenges. FEMS Microbiol. Rev. 2015, 39, 234–245. [Google Scholar] [CrossRef] [Green Version]
- Stojkovic, B.; Sretenovic, S.; Dogsa, I.; Poberaj, I.; Stopar, D. Viscoelastic properties of levan-DNA mixtures important in microbial biofilm formation as determined by micro-and macrorheology. Biophys. J. 2015, 108, 758–765. [Google Scholar] [CrossRef]
- Kesel, S.; Grumbein, S.; Gumperlein, I.; Tallawi, M.; Marel, A.K.; Lieleg, O.; Opitz, M. Direct comparison of physical properties of Bacillus subtilis NCIB 3610 and B-1 Biofilms. Appl. Environ. Microbiol. 2016, 82, 2424–2432. [Google Scholar] [CrossRef]
- Di Stefano, A.; D’Aurizio, E.; Trubiani, O.; Grande, R.; Di Campli, E.; Di Giulio, M.; Di Bartolomeo, S.; Sozio, P.; Iannitelli, A.; Nostro, A.; et al. Viscoelastic properties of Staphylococcus aureus and Staphylococcus epidermidis mono-microbial biofilms. Microb. Biotechnol. 2009, 2, 634–641. [Google Scholar] [CrossRef]
- Pavlovsky, L.; Younger, J.G.; Solomon, M.J. In situ rheology of Staphylococcus epidermidis bacterial biofilms. Soft Matter 2013, 9, 122–131. [Google Scholar] [CrossRef]
- Grumbein, S.; Werb, M.; Opitz, M.; Lieleg, O. Elongational rheology of bacterial biofilms in situ. J. Rheol. 2016, 60, 1085–1094. [Google Scholar] [CrossRef]
- Galy, O.; Latour-Lambert, P.; Zrelli, K.; Ghigo, J.M.; Beloin, C.; Henry, N. Mapping of bacterial biofilm local mechanics by magnetic microparticle actuation. Biophys. J. 2012, 103, 1400–1408. [Google Scholar] [CrossRef]
- Cao, H.Y.; Habimana, O.; Safari, A.; Heffernan, R.; Dai, Y.H.; Casey, E. Revealing region-specific biofilm viscoelastic properties by means of a micro-rheological approach. NPJ Biofilms Microbiomes 2016, 2, 5. [Google Scholar] [CrossRef]
- Chew, S.C.; Kundukad, B.; Seviour, T.; van der Maarel, J.R.C.; Yang, L.; Rice, S.A.; Doyle, P.; Kjelleberg, S. Dynamic remodeling of microbial biofilms by functionally distinct exopolysaccharides. Mbio 2014, 5, e01536-14. [Google Scholar] [CrossRef]
- Olofsson, A.C.; Hermansson, M.; Elwing, H. Use of a quartz crystal microbalance to investigate the antiadhesive potential of N-acetyl-L-cysteine. Appl. Environ. Microbiol. 2005, 71, 2705–2712. [Google Scholar] [CrossRef]
- Chen, J.Y.; Penn, L.S.; Xi, J. Quartz crystal microbalance: Sensing cell-substrate adhesion and beyond. Biosens. Bioelectron. 2018, 99, 593–602. [Google Scholar] [CrossRef]
- Dixon, M.C. Quartz crystal microbalance with dissipation monitoring: Enabling real-time characterization of biological materials and their interactions. J. Biomol. Tech. 2008, 19, 151–158. [Google Scholar]
- Costa, F.; Sousa, D.M.; Parreira, P.; Lamghari, M.; Gomes, P.; Martins, M.C.L. N-acetylcysteine-functionalized coating avoids bacterial adhesion and biofilm formation. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef]
- Tonda-Turo, C.; Carmagnola, I.; Ciardelli, G. Quartz crystal microbalance with dissipation monitoring: A powerful method to predict the in vivo behavior of bioengineered surfaces. Front. Bioeng. Biotechnol. 2018, 6, 17374. [Google Scholar] [CrossRef]
- Reipa, V.; Almeida, J.; Cole, K.D. Long-term monitoring of biofilm growth and disinfection using a quartz crystal microbalance and reflectance measurements. J. Microbiol. Methods 2006, 66, 449–459. [Google Scholar] [CrossRef]
- Sprung, C.; Wahlisch, D.; Huttl, R.; Seidel, J.; Meyer, A.; Wolf, G. Detection and monitoring of biofilm formation in water treatment systems by quartz crystal microbalance sensors. Water Sci. Technol. 2009, 59, 543–548. [Google Scholar] [CrossRef]
- Wang, Y.N.; Narain, R.; Liu, Y. Study of bacterial adhesion on different glycopolymer surfaces by quartz crystal microbalance with dissipation. Langmuir 2014, 30, 7377–7387. [Google Scholar] [CrossRef]
- Knowles, B.R.; Yang, D.; Wagner, P.; Maclaughlin, S.; Higgins, M.J.; Molino, P.J. Zwitterion functionalized silica nanoparticle coatings: The effect of particle size on protein, bacteria, and fungal spore adhesion. Langmuir 2019, 35, 1335–1345. [Google Scholar] [CrossRef]
- Tam, K.; Kinsinger, N.; Ayala, P.; Qi, F.; Shi, W.; Myung, N.V. Real-time monitoring of Streptococcus mutans biofilm formation using a quartz crystal microbalance. Caries Res. 2007, 41, 474–483. [Google Scholar] [CrossRef]
- Olsson, A.L.; van der Mei, H.C.; Busscher, H.J.; Sharma, P.K. Influence of cell surface appendages on the bacterium-substratum interface measured real-time using QCM-D. Langmuir 2009, 25, 1627–1632. [Google Scholar] [CrossRef]
- Pranzetti, A.; Salaun, S.; Mieszkin, S.; Callow, M.E.; Callow, J.A.; Preece, J.A.; Mendes, P.M. Model organic surfaces to probe marine bacterial adhesion kinetics by surface plasmon resonance. Adv. Funct. Mater. 2012, 22, 3672–3681. [Google Scholar] [CrossRef]
- Zhang, P.; Guo, J.S.; Yan, P.; Chen, Y.P.; Wang, W.; Dai, Y.Z.; Fang, F.; Wang, G.X.; Shen, Y. Dynamic dispersal of surface layer biofilm induced by nanosized tio2 based on surface plasmon resonance and waveguide. Appl. Environ. Microbiol. 2018, 84, e00047-18. [Google Scholar] [CrossRef]
- Gordon, P.W.; Brooker, A.D.M.; Chew, Y.M.J.; Wilson, D.I.; York, D.W. A scanning fluid dynamic gauging technique for probing surface layers. Meas. Sci. Technol. 2010, 21, 085103. [Google Scholar] [CrossRef]
- Peck, O.P.W.; Chew, Y.M.J.; Bird, M.R.; Bolhuis, A. Application of fluid dynamic gauging in the characterization and removal of biofouling deposits. Heat Transf. Eng. 2015, 36, 685–694. [Google Scholar] [CrossRef]
- Beyenal, H.; Babauta, J. Microsensors and microscale gradients in biofilms. Adv. Biochem. Eng. Biotechnol. 2014, 146, 235–256. [Google Scholar] [CrossRef]
- Lee, J.H.; Seo, Y.; Lim, T.S.; Bishop, P.L.; Papautsky, I. MEMS needle-type sensor array for in situ measurements of dissolved oxygen and redox potential. Environ. Sci. Technol. 2007, 41, 7857–7863. [Google Scholar] [CrossRef]
- Moya, A.; Guimera, X.; del Campo, F.J.; Prats-Alfonso, E.; Dorado, A.D.; Baeza, M.; Villa, R.; Gabriel, D.; Gamisans, X.; Gabriel, G. Biofilm oxygen profiling using an array of microelectrodes on a micro fabricated needle. Procedia Eng. 2014, 87, 256–259. [Google Scholar] [CrossRef]
- Masi, E.; Ciszak, M.; Santopolo, L.; Frascella, A.; Giovannetti, L.; Marchi, E.; Viti, C.; Mancuso, S. Electrical spiking in bacterial biofilms. J. R. Soc. Interface 2015, 12, 20141036. [Google Scholar] [CrossRef]
- James, G.A.; Zhao, A.G.; Usui, M.; Underwood, R.A.; Nguyen, H.; Beyenal, H.; Pulcini, E.D.; Hunt, A.A.; Bernstein, H.C.; Fleckman, P.; et al. Microsensor and transcriptomic signatures of oxygen depletion in biofilms associated with chronic wounds. Wound Repair Regen. 2016, 24, 373–383. [Google Scholar] [CrossRef]
- Ito, T.; Okabe, S.; Satoh, H.; Watanabe, Y. Successional development of sulfate-reducing bacterial populations and their activities in a wastewater biofilm growing under microaerophilic conditions. Appl. Environ. Microbiol. 2002, 68, 1392–1402. [Google Scholar] [CrossRef]
- Hibiya, K.; Terada, A.; Tsuneda, S.; Hirata, A. Simultaneous nitrification and denitrification by controlling vertical and horizontal microenvironment in a membrane-aerated biofilm reactor. J. Biotechnol. 2003, 100, 23–32. [Google Scholar] [CrossRef]
- Lee, W.H.; Wahman, D.G.; Pressman, J.G. Amperometric carbon fiber nitrite microsensor for in situ biofilm monitoring. Sens. Actuators B Chem. 2013, 188, 1263–1269. [Google Scholar] [CrossRef]
- von der Schulenburg, D.A.G.; Pintelon, T.R.R.; Picioreanu, C.; Van Loosdrecht, M.C.M.; Johns, M.L. Three-dimensional simulations of biofilm growth in porous media. AIChE J. 2009, 55, 494–504. [Google Scholar] [CrossRef]
- Bottero, S.; Storck, T.; Heimovaara, T.J.; van Loosdrecht, M.C.M.; Enzien, M.V.; Picioreanu, C. Biofilm development and the dynamics of preferential flow paths in porous media. Biofouling 2013, 29, 1069–1086. [Google Scholar] [CrossRef]
- Davit, Y.; Byrne, H.; Osborne, J.; Pitt-Francis, J.; Gavaghan, D.; Quintard, M. Hydrodynamic dispersion within porous biofilms. Phys. Rev. 2013, 87, 012718. [Google Scholar] [CrossRef] [Green Version]
- Qin, C.Z.; Hassanizadeh, S.M. Pore-network modeling of solute transport and biofilm growth in porous media. Transp. Porous Med. 2015, 110, 345–367. [Google Scholar] [CrossRef]
- McLean, J.S.; Ona, O.N.; Majors, P.D. Correlated biofilm imaging, transport and metabolism measurements via combined nuclear magnetic resonance and confocal microscopy. ISME J. 2008, 2, 121–131. [Google Scholar] [CrossRef]
- Phoenix, V.R.; Holmes, W.M. Magnetic resonance imaging of structure, diffusivity, and copper immobilization in a phototrophic biofilm. Appl. Environ. Microbiol. 2008, 74, 7454. [Google Scholar] [CrossRef]
- Vogt, M.; Flemming, H.C.; Veeman, W.S. Diffusion in Pseudomonas aeruginosa biofilms: A pulsed field gradient NMR study. J. Biotechnol. 2000, 77, 137–146. [Google Scholar] [CrossRef]
- Gabrilska, R.A.; Rumbaugh, K.P. Biofilm models of polymicrobial infection. Future Microbiol. 2015, 10, 1997–2015. [Google Scholar] [CrossRef] [Green Version]
- Brann, M.; Suter, J.D.; Addleman, R.S.; Larimer, C. Monitoring bacterial biofilms with a microfluidic flow chip designed for imaging with white-light interferometry. Biomicrofluidics 2017, 11, 044113. [Google Scholar] [CrossRef]
- Lourenco, A.; Coenye, T.; Goeres, D.M.; Donelli, G.; Azevedo, A.S.; Ceri, H.; Coelho, F.L.; Flemming, H.C.; Juhna, T.; Lopes, S.P.; et al. Minimum information about a biofilm experiment ( MIABiE): Standards for reporting experiments and data on sessile microbial communities living at interfaces. Pathog. Dis. 2014, 70, 250–256. [Google Scholar] [CrossRef]
- Lourenco, A.; Ferreira, A.; Veiga, N.; Machado, I.; Pereira, M.O.; Azevedo, N.F. BiofOmics: A Web platform for the systematic and standardized collection of high-throughput biofilm data. PLoS ONE 2012, 7, e39960. [Google Scholar] [CrossRef]
- Coenye, T.; Goeres, D.; Van Bambeke, F.; Bjarnsholt, T. Should standardized susceptibility testing for microbial biofilms be introduced in clinical practice? Clin. Microbiol. Infect. 2018, 24, 570–572. [Google Scholar] [CrossRef] [Green Version]






| Apparatus | Flow Dynamic | Interface | Biofilm Analysis | Advantage | Limitations | Standard Methods |
|---|---|---|---|---|---|---|
| Microtiter plate | Static | Solid–liquid | - Adhesion - Detachment - Susceptibility to antimicrobial | - Easy to use - Cheap - Reproducible - Simple equipment - High-throughput screening - Multiple materials tested simultaneously - Multiple conditions tested simultaneously - Well-designed to investigate early stage of biofilm formation - Comparable experiments among different laboratories | - Not suitable to study late stage of biofilm formation - Sensitive to cell sedimentation - Poor reproducibility of real environment - Close system - Washing step can remove not loosely attached cells - Designed to study biofilms only at solid-liquid interface | None |
| Calgary biofilm device | Static | Solid–liquid | - Adhesion - Detachment - Susceptibility to antimicrobial | - Easy to use - Cheap - Simple equipment - Reproducible - High-throughput screening - Multiple materials tested simultaneously - Multiple conditions tested simultaneously - Well-designed to investigate early stage of biofilm formation - Reduced interference of cell sedimentation - Comparable experiments among different laboratories | - Not suitable to study late stage of biofilm formation - Poor reproducibility of real environment - Close system - Washing step can remove not loosely attached cells - Designed to study biofilms only at solid-liquid interface - Difficulty to collect individual pegs for cell enumeration | ASTM E2799-17 Standard test method for testing disinfectantefficacy against Pseudomonas aeruginosa biofilm using the MBEC assay [104] |
| Biofilm ring test | Static | Solid–liquid | - Adhesion - Detachment - Susceptibility to antimicrobial | - Easy to use - Reproducible - Rapid - High-throughput screening - Automatic analysis - Very low standard deviation - No need for staining or washing steps - Multiple conditions tested simultaneously - Comparable experiments among different laboratories - Well-designed to investigate early stage of biofilm formation | - Not suitable to study late stage of biofilm formation - Poor reproducibility of real environment - Close system - Expensive - Sensitive to cell sedimentation - Only suitable for thick biofilms - Requires specific magnetic device and scanner - Need of specified software and biofilm index to provide the results - Not suitable for multi-species biofilm - Designed to study biofilms only at solid-liquid interface - the material must display magnetic properties - Not suitable to compare multiple materials simultaneously | None |
| Real Time xCelligence | Static | Solid–liquid | - Adhesion - Detachment - Susceptibility to antimicrobial | - Easy to use - Reproducible - High-throughput screening - Non-invasive - automatic analysis - Very low standard deviation - No need for staining or washing steps - Multiple conditions tested simultaneously - Comparable experiments among different laboratories - Well-designed to investigate early stage of biofilm formation | - Not suitable to study late stage of biofilm formation - Poor reproducibility of real environment - Close system - Expensive - Sensitive to cell sedimentation - Requires specific equipment - Not suitable for multi-species biofilm - Designed to study biofilms only at solid-liquid interface - Not suitable to compare multiple materials simultaneously - Materials must have electrical properties | None |
| Colony biofilm | Static | Solid–air | - Adhesion - Susceptibility to antimicrobial | - Easy to use - Cheap - Simple equipment - Reproducible - Large biomass in a short period | - Planktonic cells may interfere with the biofilm assays - Biofilm detachment can not be studied - Material must be customized only in the form of semipermeable membrane-like thin film - Difficulty in manipulating surfaces when colony biomass become large - Designed to study biofilms only at solid-air interface | None |
| Transwell | Static | Solid–air | - Adhesion - Susceptibility to antimicrobial | - Easy to use - Cheap - Simple equipment - Reproducible - High-throughput screening - Large biomass in a short period - Possibility to collect the metabolites from biofilm culture - Independent studies in compartments with different condition | - Biofilm detachment can not be studied - Material must be customized only in the form of semipermeable membrane-like thin film - Difficulty in manipulating surfaces when colony biomass become large - Designed to study biofilms only at air-solid interface | None |
| Robbins Device (RD) | Dynamic (variable laminar/turbulent shear) | Solid–liquid | - Adhesion - Maturation - Detachment - Susceptibility to antimicrobial | - Multiple materials tested simultaneously (up to 12) - Suitable to study early and late stage of biofilm formation - Reproducibility of real environments - Large biomass in a short period - Can operate under differenthydrodynamic conditions (e.g., laminar/turbulent) - Autoclavable and re-useable | - Not suitable for high-throughput screening - Necessity of specialized equipment - Technically challenging - Poor experimental reproducibility - Multiple microorganisms can not be tested simultaneously - Designed to study biofilms only at solid-liquid interface - No direct observation of biofilm development - Low air-liquid exchange - Flow inside the device is difficult to be accurately adjusted - Fixed geometry of the surface (disk) - Expensive - Requires specific equipment | None |
| Center for Disease Control (CDC) reactor | Dynamic (high laminar/turbulent shear) | Solid–liquid | - Adhesion - Maturation - Detachment - Susceptibility to antimicrobial | - Multiple materials tested simultaneously (up to 24) - Suitable to study early and late stage of biofilm formation - Reproducibility of real environments - Large biomass in a short period - Well controlled flow within the device - All surface within the device experience the share stress - Suitable for time-course studies - Can operate under differenthydrodynamic conditions (e.g., low/high and laminar/turbulent share) - Standardized biofilm method - Autoclavable and re-useable | - Not suitable for high-throughput screening - Requires specific equipment - Technically challenging - Multiple microorganisms can not be tested simultaneously - Low air-liquid exchange - Designed to study biofilms only at solid-liquid interface - No direct observation of biofilm development - Fixed geometry of the surface (disk) - Expensive - Requires a large volume of medium | - ASTM E2562-17 Standard test method for quantification of Pseudomonas aeruginosa biofilm grown with high shear and continuous flow using CDC biofilm reactor [105] - ASTM E2871-19 Standard test method for determining disinfectant efficacy against biofilm grown in the CDC biofilm reactor using the single tube method [106] |
| Rotating Disk (RD) reactor | Dynamic (intermediate laminar/turbulent shear) | Solid–liquid | - Adhesion - Maturation - Detachment - Susceptibility to antimicrobial | - Multiple materials tested simultaneously (up to 6) - Suitable to study early and late stage of biofilm formation - Reproducibility of real environments - Large biomass in a short period - Different shear can be tested simultaneously - Standardized biofilm method - Autoclavable and re-useable | - Not suitable for high-throughput screening - Requires specific equipment - Technically challenging - Multiple microorganisms can not be tested simultaneously - Low air–liquid exchange - Designed to study biofilms only at solid-liquid interface - Less suitable for time-course studies - No direct observation of biofilm development - Fixed geometry of the surface (disk) - Requires a large volume of medium - Expensive | ASTM E2196-17 Standard test method for quantification of a Pseudomonas aeruginosa biofilm grown with shear and continuous flow using a rotating disk reactor [107] |
| Annular (RA) reactor | Dynamic (variable laminar/turbulent shear) | Solid–liquid | - Adhesion - Maturation - Detachment - Susceptibility to antimicrobial | - Multiple materials tested simultaneously (up to 20) - Suitable to study early and late stage of biofilm formation - Reproducibility of real environments - Large biomass in a short period - Suitable for time-course studies - Can operate under differenthydrodynamic conditions (e.g., low/high and laminar/turbulent share) - Autoclavable and re-useable | - Not suitable for high-throughput screening - Requires specific equipment - Technically challenging - Multiple microorganisms can not be tested simultaneously - Low air-liquid exchange - Designed to study biofilms only at solid-liquid interface - No direct observation of biofilm development - Fixed geometry of the surface (rectangular) - Requires a large volume of medium - Expensive | None |
| Drip flow (DF) reactor | Dynamic (low laminar shear) | Solid–air | - Adhesion - Maturation - Detachment - Susceptibility to antimicrobial | - Multiple materials tested simultaneously (up to 4 or 6) - Suitable to study early and late stage of biofilm formation - Reproducibility of real environments - Large biomass in a short period - Suitable for time-course studies - Standardized biofilm method - Multiple microorganisms can be tested simultaneously - Autoclavable and re-useable - Compatible with surface of variousgeometry - High gas transfer | - Heterogeneity of biofilm developmenton the coupons - Not suitable for high-throughput screening - Requires specific equipment - Technically challenging - Designed to study biofilms only at solid-liquid interface - Direct observation of the biofilm development is not allowed - Expensive | ASTM E2647-13 Standard test method for quantification of Pseudomonas aeruginosa biofilm grown using drip flow biofilm reactor with low shear and continuous flow [108] |
| Flow chamber | Dynamic (variable laminar/turbulent shear) | Solid–liquid | - Adhesion - Maturation - Detachment - Susceptibility to antimicrobial | - Multiple materials tested simultaneously (up to 6) - Suitable to study early and late stage of biofilm formation - Reproducibility of real environments - Direct inspection of living biofilm development in a non-destructive way - Multiple microorganisms can be tested simultaneously - Autoclavable and re-useable | - Low biomass - Not suitable for high-throughput screening - Requires specific equipment - Technically challenging - Designed to study biofilms only at solid-liquid interface - Biofilm recovery during experiment is difficult - Expensive - Problem of air bubble | None |
| Microplate under flow (Bioflux) | Dynamic (variable laminar shear) | Solid–liquid | - Adhesion - Maturation - Detachment - Susceptibility to antimicrobial | - Easy to use - Cheap - Reproducible - Simple equipment - High-throughput screening - Multiple materials tested simultaneously (up to 96) - Multiple conditions tested simultaneously - Suitable to study early and late stage of biofilm formation - Reproducibility of real environments - Direct inspection of living biofilm development in a non-destructive way - Suitable for time-course studies - Multiple microorganisms can be tested simultaneously | - Low biomass - Designed to study biofilms only at solid-liquid interface - No biofilm recovery during experiment | |
| Microcosm | Static, Dynamic (variable laminar/turbulent shear) | Solid–liquid, solid–air | - Adhesion - Maturation - Detachment - Susceptibility to antimicrobial | - Hight reproducibility of real environments - Multiple materials tested simultaneously - Suitable to study early and late stage of biofilm formation - Multiple microorganisms can be tested simultaneously - Both static and dynamic systems - Compatible with surface of variousgeometry | - Less reproducibility - Not suitable for high-throughput screening - Technically challenging - Biofilm recovery during experiment is difficult - Complicated interpretation of results - Not suitable for time-course studies - No direct observation of biofilm development | None |
| Topic | Method | In Situ | Ex Situ | Surface Chemistry Modification | Surface Topography Modification | Surface with Microstructure | Antimicrobial Coating | Antimicrobial-Releasing Surface | Antimicrobial-Responsive Surface | Covalent Immobilization of Antimicrobials | Metal Coating | Nanoparticles Based Materials | Anti-Biofilm Biocide-Free Surfaces |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Viable cellular biomass | Plate count assay | ✖ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ |
| Biomarkers quantification (Phospholipid fatty acids, ergosterol) | ✖ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | |
| Metabolic colorimetric dyes | ✔ | ✖ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | |
| ATP bioluminescence | ✖ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | |
| Isothermal microcalorimetry | ✔ | ✖ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | |
| Tunable Diode Laser Absorption Spectroscopy | ✔ | ✖ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | |
| Propidium monoazide-qPCR | ✖ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | |
| Total (viable and not viable) cellular biomass | Chamber counting | ✖ | ✔ | ✔ | ✔ | ✖ | ✖ | ✖ | ✖ | ✖ | ✖ | ✔ | ✔ |
| Dyes binding | ✔ | ✖ | ✔ | ✔ | ✖ | ✖ | ✖ | ✖ | ✖ | ✖ | ✔ | ✔ | |
| Biomarker quantification (total organic carbon, proteins, chlorophyll) | ✖ | ✔ | ✔ | ✔ | ✖ | ✖ | ✖ | ✖ | ✖ | ✖ | ✔ | ✔ | |
| Quantitative polymerase chain reaction (q-PCR) | ✖ | ✔ | ✔ | ✔ | ✖ | ✖ | ✖ | ✖ | ✖ | ✖ | ✔ | ✔ | |
| Flow-based cell counting | ✖ | ✔ | ✔ | ✔ | ✖ | ✖ | ✖ | ✖ | ✖ | ✖ | ✔ | ✔ | |
| Total biofilm (cellular biomass + EPS) | Dry weight | ✔ | ✖ | ✔ | ✔ | ✖ | ✖ | ✖ | ✖ | ✖ | ✖ | ✔ | ✔ |
| Optical density | ✖ | ✔ | ✔ | ✔ | ✖ | ✖ | ✖ | ✖ | ✖ | ✖ | ✔ | ✔ | |
| Dye-based methods | ✔ | ✖ | ✔ | ✔ | ✖ | ✖ | ✖ | ✖ | ✖ | ✖ | ✔ | ✔ | |
| Colour measurements | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | |
| EPS matrix | Proteins quantification | ✖ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ |
| Polysaccharides quantification | ✖ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | |
| Extracellular DNA quantification | ✖ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | |
| Antibody microarrays | ✖ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | |
| Fluorescent microscopies | ✔ | ✖ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | |
| Spectroscopic techniques (ATR-IR, Raman, SERS) | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | |
| Advanced techniques (HPLC, LC–UV-ESI-MS/MS, GC/MS, NMR, TXRF, ICP-MS) | ✖ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | |
| Identification and spatial distribution of biofilm members | FISH and FISH-based techniques | ✔ | ✖ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ |
| Immunofluorescence detection | ✔ | ✖ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | |
| Biofilm morphology | Confocal Laser Scanning Microscopy (CLSM, FRAP, FCS, FLIM) | ✔ | ✖ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ |
| Multiphoton microscopy (MPM) | ✔ | ✖ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | |
| Electronic microscopies (SEM, TEM, cryo-SEM, ESEM, FIB-SEM, ASEM) | ✔ | ✖ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | |
| Raman microscopy (RM, CRM) | ✔ | ✖ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | |
| Scanning transmission X-ray microscopy (STXM) | ✔ | ✖ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | |
| Super-resolution microscopies (STED, GSD, RESOLFT, PALM, STORM, SIM, SSIM) | ✔ | ✖ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | |
| Optical coherence tomography (OCT) | ✔ | ✖ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | |
| Mechanical and physical properties | Atomic force microscopy (AFM, AFM-SCFS) | ✔ | ✖ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ |
| Rheometry (macro and micro-reometry, SPT, MPT) | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | |
| Quartz crystal microbalance (QCM, QCM-D) | ✔ | ✖ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | |
| Surface plasma resonance (SPR) | ✔ | ✖ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | |
| Fluid dynamic gauging (FDG) | ✔ | ✖ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | |
| Microsensors | ✔ | ✖ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | |
| Nuclear magnetic resonance (NMR, MRI, PFG-NMR) | ✔ | ✖ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ |
| Staining Pattern | Stain | Colour (Maxima Excitation/ Emission) | Cells (Viability: Live/Dead/ Both) | Matrix | Polymeric Substrate |
|---|---|---|---|---|---|
| Nucleic acids | DAPI | Blue (358/461 nm) | X (both) | - Polyurethane coated with N-vanillylnonanamide [292] - Permanox plastic coated with B-type proanthocyanidins [293] - Surfactant polymer dressing (PluroGel) [294] | |
| SYTO 9 | Green (485/498 nm) | X (both) | - Poly(ε-caprolactone-co-δ-valerolactone) mixed with dibromohemibasta-din-1 [81] - Silycon containing (3-acrylamidopropyl) trimethylammonium chloride (AMPTMA) or quaternized polyethylenimine methacrylate [99] - Siloxane modified with non-ionic surfactants and antioxidant [295] | ||
| SYBR green-I | Green (495/537 nm) | X (both) | - Low density polyethylene functionalyzed with p-aminocinnamic acid and p-aminosalicylic acid [284] - Low density polyethylene functionalized with α-chymotrypsin [74] | ||
| Live/Dead Bac-Light | Green (485/498 nm) Red (585/617 nm) | X (both) | - Nanostructured silicon [55] - Black silicon with nanoprotrusion [296] - Nanoporous 1,2-polybutadiene-b-polydimethylsiloxane loaded with sodium dodecyl sulfate [297] - Nanostructured poly (methyl methacrylate) [298] - Polydimethylsiloxane with immobilized polymyxins B and E [28] - NO-releasing amine plasma polymer [299] - Low density polyethylene functionalyzed with p-aminocinnamic acid and p-aminosalicylic acid [284] - Low density polyethylene functionalized with α-chymotrypsin [74] - Poly(methyl methacrylate) with incorporated Juglone [113] - Ciprofloxacin-incorporated polyurethane polymers [300] | ||
| Propidium Iodide | Red (585/617 nm) | X (dead) | - Nanoporous 1,2-polybutadiene-b-polydimethylsiloxane leaded with sodium dodecyl sulfate [297] - Polymer brushes based on poly(cysteine methacrylate) grafted on on the nanocellulose membranes [301] | ||
| Acridine orange | Green (500/526 nm) | X (both) | - Urinary catheters coated with lipase-embedded polycaprolactone (PCL), coimpregnated with the antibiotic gentamicin sulfate [302] | ||
| Plasma membrane | CellMask plasma membrane orange | Red (554/567 nm) | X (both) | - Low density polyethylene functionalyzed with p-aminocinnamic acid and p-aminosalicylic acid [76] | |
| Cell wall | Wheat Germ Agglutinin (FITC conjugated) | Green (490/525 nm) | X (both) | - Surfactant polymer dressing (PluroGel) [294] | |
| Calcofluor white | Blue (355/433 nm) | X (both) | - 1H,1H,2H,2H-Perfluoro-octyl-methacrylate, methyl methacrylate, Paraloid B72 acrylic resin [303] | ||
| Metabolic activity | Resazurin | Blu (575/585 nm) | X (live) | - Poly(vinyl alcohol-co-ethylene) [101] | |
| Calcein or fluorescein diacetate | Green (495/515 nm) | X (live) | - Poly(vinyl alcohol-co-ethylene) [101] | ||
| SNARF-1 | Green (580/640 nm) | X | - Poly(methacrylic acid) antibiotic-loaded [304] | ||
| Proteins | FITC | Green (490/525 nm) | X (both) | - Poly (ε-caprolactone)-based polyurethane coated with butanolide [53] | |
| SYPRO Ruby | Red (450/610 nm) | X | - Surfactant polymer dressing (PluroGel) [294] - Polymer brushes based on poly(cysteine methacrylate) grafted on on the nanocellulose membranes [301] | ||
| Polysaccharides | Concanavalin A (Texas red, Alexa Fluor 594 conjugated) | Red (Texas red: 595/613 nm; Alexa Fluor 594: 591/618 nm;) | X (both) | X | - Low density polyethylene functionalyzed with p-aminocinnamic acid and p-aminosalicylic acid [284] - Low density polyethylene functionalized with α-chymotrypsin [74] - Silica particles attached to polydimethylsiloxane [305] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cattò, C.; Cappitelli, F. Testing Anti-Biofilm Polymeric Surfaces: Where to Start? Int. J. Mol. Sci. 2019, 20, 3794. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20153794
Cattò C, Cappitelli F. Testing Anti-Biofilm Polymeric Surfaces: Where to Start? International Journal of Molecular Sciences. 2019; 20(15):3794. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20153794
Chicago/Turabian StyleCattò, Cristina, and Francesca Cappitelli. 2019. "Testing Anti-Biofilm Polymeric Surfaces: Where to Start?" International Journal of Molecular Sciences 20, no. 15: 3794. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20153794