Mouse Tryptase Gene Expression is Coordinately Regulated by GATA1 and GATA2 in Bone Marrow-Derived Mast Cells
Abstract
:1. Introduction
2. Results
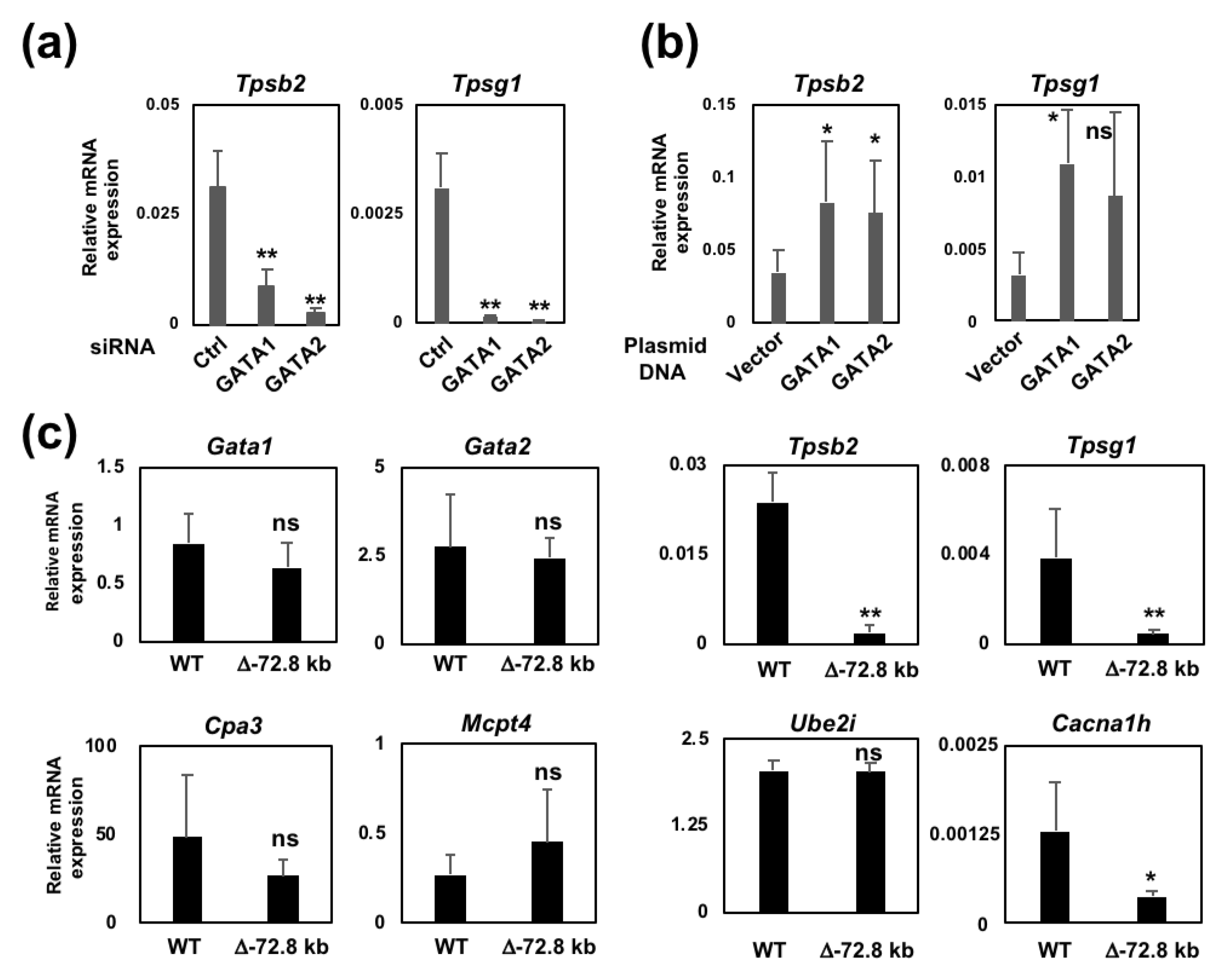
2.1. The Introduction of siRNA Targeting Either GATA1 or GATA2 into BMMCs Leads to a Significant Reduction in Mast Cell Tryptase Gene Expression
2.2. Overexpression of MITF Failed to Restore the Reduced Tpsb2 Transcript Level in the GATA1/GATA2 Double-Knockdown BMMCs
2.3. GATA1 and GATA2 Bind to Three Genomic Regions Upstream of the Tpsb2 Gene
2.4. The GATA2 Binding Activity to the −72.8 kb Region is Reduced by GATA1 Ablation
2.5. The −72.8 kb Upstream Region of the Tpsb2 Gene is Indispensable for the Expression of the Tpsb2 and Tpsg1 Genes in MEDMC-BRC6 Murine Mast Cells
2.6. GATA1 Regulates the CTCF and Rad21 Binding Activity to the Tryptase Gene Locus
3. Discussion
4. Materials and Methods
4.1. Mice
4.2. Cell Culture
4.3. Transfection of Small Interfering RNA (siRNA) or Plasmid DNA
4.4. Quantitative RT-PCR (qRT-PCR)
4.5. Western Blotting
4.6. Chromatin Immunoprecipitation (ChIP)
4.7. CRISPR/Cas9 Genome Editing
4.8. ChIP-seq Data Processing
4.9. Statistical Analyses
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| BMMCs | Bone marrow-derived mast cells |
| ChIP | Chromatin immunoprecipitation |
| MITF | Microphthalmia-associate transcription factor |
| RT | Reverse transcription |
| qPCR | Quantitative polymerase chain reaction |
| RNA-seq | RNA sequencing |
| RPKM | reads per kilobase million |
| ChIP-seq | chromatin immunoprecipitation sequencing |
| 3C | chromosome conformation capture |
| TAD | topologically associating domains |
| 4-OHT | 4-hydroxy tamoxifen |
| RPMI | Roswell Park Memorial Institute |
| IMDM | Iscove’s modified Dulbecco’s medium |
| FBS | fetal bovine serum |
| Polr2a | polymerase (RNA) II (DNA directed) polypeptide A |
| GFP | green fluorescent protein |
| sgRNA | single-guide RNA |
| IGV | Integrative Genomics Viewer |
Appendix A
| Primers for qRT-PCR | Sequences (5′ to 3′) | |
|---|---|---|
| Forward | Reverse | |
| Gata1 | CAGAACCGGCCTCTCATCC | TAGTGCATTGGGTGCCTGC |
| Gata2 | GCACCTGTTGTGCAAATTGT | GCCCCTTTCTTGCTCTTCTT |
| Cebpa | AAAGCCAAGAAGTCGGTGGAC | CTTTATCTCGGCTCTTGCGC |
| Mitf | GCTGGAGATGCAGGCTAGAG | TGATGATCCGATTCACCAGA |
| Cpa3 | GCTACACATTCAAACTGCCTCCT | GAGAGAGCATCCGTGGCAA |
| Mcpt4 | CATGCTTTGTTGAACCCAAGG | GAAGTGAAAAGCCTGACCTGC |
| Mcpt8 | GTGGGAAATCCCAGTG | GACAACCATACCCCAG |
| Cma1 | CCTGGGTTCCAGCACCAA | GGCGGGAGTGTGGTATGC |
| Ctsg | GAGTCCAGAAGGGGCTG | GATGGCTCTGAGACAT |
| Tpsb2 | CGACATTGATAATGACGAGCCTC | ACAGGCTGTTTTCCACAATGG |
| Tpsg1 | GGTCACACTGTCTCCCCACT | GCATCCCAGGGTAGAAGTCA |
| Mitf ex1a | AAGTCGGGGAGGAGTTTCAC | CATCAATTACATCATCCATCTGCATGC |
| Mitf ex1mc | CGACAAGCTTATGAACCGGCTTTTCCTG | |
| Mitf ex1e | TCACAGAGGTTAGTAGGTGGATGGG | |
| Mitf ex1h | GGCGCTTAGATTTGAGATGC | |
| Mitf ex1m | GAGGACTAAGTGGTCTGCGG | |
| Mitf ex1b | CCTGAGCTCACCATGTCCAAAC | |
| Prss34 | GCTGATGAAAGTGGTCAAGATCATCCG | AGGAGTGAATGCATCAATATGAGTGGCTG |
| Prss28 | GTACCGTGTTCATGGCCTCT | TGACTTTGGATGCAGTGAGC |
| Prss29 | GTCAAGCTGCCCTCTGAGTC | TGGTTGCCTGCACATAACAT |
| Tpsab1 | ATGACCACCTGATGACTGTGAGCCAG | AGGAACGGAGGTCATCCTGGATGTG |
| Ube2i | GAGGCTTGTTCAAGCTACGG | GTGATAGCTGGCCTCCAGTC |
| Cacna1h | CCTTTCTCAGCGTCTCCAAC | GCCACAATGATGTCAACCAG |
| Polr2a | CTGGACCCTCAAGCCCATACAT | CGTGGCTCATAGGCTGGTGAT |
| Primers | Sequences (5′ to 3′) | ||
|---|---|---|---|
| Forward | Reverse | ||
| Tpsb2 | −72.8 kb | AACCTTCGACGTGACCTTTG | GGCACAGGATTTGTGAGACC |
| −67.9 kb | TCAGTTGGCAGGTTTCTGTG | ACCAGTCAGGGCAAGTTCAC | |
| −63.4 kb | CTTACTGCTTTGGCCTGGAG | TATGAATTTGGAGGCGATCC | |
| −34.0 kb | AGCCTCCTAAGGGTCAGAGC | CCGCGATATTATCTGCACCT | |
| −1.1 kb | GGAGGTCACACTGCAGGATT | AATGGGTAACAGCGTCGTTC | |
| +2.6 kb | CTTGCCTTCCTTGTCCTCTG | AGAAGAGAGGGAGCCACACA | |
| −81.3 kb | GTGAGGCCCATTCAAAAGAA | CTAGGGAACCAGATGCCAAA | |
| −75.9 kb | GAGGACCCAAGTGAGGTTCA | AGAAGCCCCTAGGAGTGAGC | |
| −2.4 kb | CAGTCCAACTGCACCAACC | CACCCTCAGTTGCCTCCTCA | |
| Gata2 | −3.9 kb | GAGATGAGCTAATCCCGCTGTA | AAGGCTGTATTTTTCCAGGCC |
| −27.7 kb | TGCCATGCCGGATATATTTTG | ACTAGCACGTGTGGCACAGTG | |
| Kit | −114 kb | CGTGCACACAGGTTTGTTTC | TGCTGAGATGTGGCAATAGG |
| Applied Oligonucleotide | Sequences of Oligonucleotide (5′ to 3′) | |
|---|---|---|
| sgRNAs | 5′-fwd | CACCGTCTTACTGTAGATTTAAGC |
| 5′-rev | AAACGCTTAAATCTACAGTAAGAC | |
| 3′-fwd | CACCGACATTAGAACACTATTAGTA | |
| 3′-rev | AAACTACTAATAGTGTTCTAATGTC | |
| 5′ arm | Fwd | TAATGGGCCCACAGGGCCTCATTTTGTAGG |
| Rev | ACCGATCGATGCTTAAATCTACAGTAAGAC | |
| 3′ arm | Fwd | TATGGGATCCACTACTTTTACACTGCCTCA |
| Rev | ACTTGCGGCCGCTACATGTTGGCACAAGCCTG | |
References
- Caughey, G.H. Tryptase Genetics and Anaphylaxis. J. Allergy Clin. Immunol. 2006, 117, 1411–1414. [Google Scholar] [PubMed]
- Akin, C.; Soto, D.; Brittain, E.; Chhabra, A.; Schwartz, L.B.; Caughey, G.H.; Metcalfe, D.D. Tryptase Haplotype in Mastocytosis: Relationship to Disease Variant and DiagnostOlic Utility of Total Tryptase Levels. Clin. Immunol. 2007, 123, 268–271. [Google Scholar] [CrossRef] [PubMed]
- Thakurdas, S.M.; Melicoff, E.; Sansores-Garcia, L.; Moreira, D.C.; Petrova, Y.; Stevens, R.L.; Adachi, R. The Mast Cell-Restricted Tryptase mMCP-6 has a Critical Immunoprotective Role in Bacterial Infections. J. Biol. Chem. 2007, 282, 20809–20815. [Google Scholar] [CrossRef] [PubMed]
- Shin, K.; Watts, G.F.; Oettgen, H.C.; Friend, D.S.; Pemberton, A.D.; Gurish, M.F.; Lee, D.M. Mouse Mast Cell Tryptase mMCP-6 is a Critical Link between Adaptive and Innate Immunity in the Chronic Phase of Trichinella Spiralis Infection. J. Immunol. 2008, 180, 4885–4891. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, N.N.; Tamraz, B.; Chu, C.; Kwok, P.Y.; Caughey, G.H. Human Subjects are Protected from Mast Cell Tryptase Deficiency Despite Frequent Inheritance of Loss-of-Function Mutations. J. Allergy Clin. Immunol. 2009, 124, 1099–1105.e4. [Google Scholar] [CrossRef]
- Trivedi, N.N.; Caughey, G.H. Mast Cell Peptidases: Chameleons of Innate Immunity and Host Defense. Am. J. Respir. Cell Mol. Biol. 2010, 42, 257–267. [Google Scholar] [CrossRef] [PubMed]
- Reimer, J.M.; Samollow, P.B.; Hellman, L. High Degree of Conservation of the Multigene Tryptase Locus Over the Past 150-200 Million Years of Mammalian Evolution. Immunogenetics 2010, 62, 369–382. [Google Scholar] [CrossRef]
- Hunt, J.E.; Stevens, R.L.; Austen, K.F.; Zhang, J.; Xia, Z.; Ghildyal, N. Natural Disruption of the Mouse Mast Cell Protease 7 Gene in the C57BL/6 Mouse. J. Biol. Chem. 1996, 271, 2851–2855. [Google Scholar] [CrossRef] [Green Version]
- Wong, G.W.; Yasuda, S.; Morokawa, N.; Li, L.; Stevens, R.L. Mouse Chromosome 17A3.3 Contains 13 Genes that Encode Functional Tryptic-Like Serine Proteases with Distinct Tissue and Cell Expression Patterns. J. Biol. Chem. 2004, 279, 2438–2452. [Google Scholar] [CrossRef]
- Lyons, J.J.; Yu, X.; Hughes, J.D.; Le, Q.T.; Jamil, A.; Bai, Y.; Ho, N.; Zhao, M.; Liu, Y.; O’Connell, M.P.; et al. Elevated Basal Serum Tryptase Identifies a Multisystem Disorder Associated with Increased TPSAB1 Copy Number. Nat. Genet. 2016, 48, 1564–1569. [Google Scholar] [CrossRef]
- Cildir, G.; Pant, H.; Lopez, A.F.; Tergaonkar, V. The Transcriptional Program, Functional Heterogeneity, and Clinical Targeting of Mast Cells. J. Exp. Med. 2017, 214, 2491–2506. [Google Scholar] [CrossRef] [PubMed]
- Morii, E.; Tsujimura, T.; Jippo, T.; Hashimoto, K.; Takebayashi, K.; Tsujino, K.; Nomura, S.; Yamamoto, M.; Kitamura, Y. Regulation of Mouse Mast Cell Protease 6 Gene Expression by Transcription Factor Encoded by the Mi Locus. Blood 1996, 88, 2488–2494. [Google Scholar] [PubMed]
- Ogihara, H.; Kanno, T.; Morii, E.; Kim, D.K.; Lee, Y.M.; Sato, M.; Kim, W.Y.; Nomura, S.; Ito, Y.; Kitamura, Y. Synergy of PEBP2/CBF with Mi Transcription Factor (MITF) for Transactivation of Mouse Mast Cell Protease 6 Gene. Oncogene 1999, 18, 4632–4639. [Google Scholar] [CrossRef] [PubMed]
- Ogihara, H.; Morii, E.; Kim, D.K.; Oboki, K.; Kitamura, Y. Inhibitory Effect of the Transcription Factor Encoded by the Mutant Mi Microphthalmia Allele on Transactivation of Mouse Mast Cell Protease 7 Gene. Blood 2001, 97, 645–651. [Google Scholar] [CrossRef] [PubMed]
- Morii, E.; Ogihara, H.; Oboki, K.; Kataoka, T.R.; Jippo, T.; Kitamura, Y. Effect of MITF on Transcription of Transmembrane Tryptase Gene in Cultured Mast Cells of Mice. Biochem. Biophys. Res. Commun. 2001, 289, 1243–1246. [Google Scholar] [CrossRef] [PubMed]
- Hu Frisk, J.M.; Kjellen, L.; Kaler, S.G.; Pejler, G.; Ohrvik, H. Copper Regulates Maturation and Expression of an MITF:Tryptase Axis in Mast Cells. J. Immunol. 2017, 199, 4132–4141. [Google Scholar] [CrossRef] [PubMed]
- Ohneda, K.; Moriguchi, T.; Ohmori, S.; Ishijima, Y.; Satoh, H.; Philipsen, S.; Yamamoto, M. Transcription Factor GATA1 is Dispensable for Mast Cell Differentiation in Adult Mice. Mol. Cell. Biol. 2014, 34, 1812–1826. [Google Scholar] [CrossRef] [PubMed]
- Ohmori, S.; Moriguchi, T.; Noguchi, Y.; Ikeda, M.; Kobayashi, K.; Tomaru, N.; Ishijima, Y.; Ohneda, O.; Yamamoto, M.; Ohneda, K. GATA2 is Critical for the Maintenance of Cellular Identity in Differentiated Mast Cells Derived from Mouse Bone Marrow. Blood 2015, 125, 3306–3315. [Google Scholar] [CrossRef] [PubMed]
- Takemoto, C.M.; Yoon, Y.J.; Fisher, D.E. The Identification and Functional Characterization of a Novel Mast Cell Isoform of the Microphthalmia-Associated Transcription Factor. J. Biol. Chem. 2002, 277, 30244–30252. [Google Scholar] [CrossRef]
- Calero-Nieto, F.J.; Ng, F.S.; Wilson, N.K.; Hannah, R.; Moignard, V.; Leal-Cervantes, A.I.; Jimenez-Madrid, I.; Diamanti, E.; Wernisch, L.; Gottgens, B. Key Regulators Control Distinct Transcriptional Programmes in Blood Progenitor and Mast Cells. EMBO J. 2014, 33, 1212–1226. [Google Scholar] [CrossRef]
- Li, Y.; Liu, B.; Harmacek, L.; Long, Z.; Liang, J.; Lukin, K.; Leach, S.M.; O’Connor, B.; Gerber, A.N.; Hagman, J.; et al. The Transcription Factors GATA2 and Microphthalmia-Associated Transcription Factor Regulate Hdc Gene Expression in Mast Cells and are Required for IgE/mast Cell-Mediated Anaphylaxis. J. Allergy Clin. Immunol. 2018, 142, 1173–1184. [Google Scholar] [CrossRef] [PubMed]
- Ohmori, S.; Takai, J.; Ishijima, Y.; Suzuki, M.; Moriguchi, T.; Philipsen, S.; Yamamoto, M.; Ohneda, K. Regulation of GATA Factor Expression is Distinct between Erythroid and Mast Cell Lineages. Mol. Cell. Biol. 2012, 32, 4742–4755. [Google Scholar] [CrossRef] [PubMed]
- Hiroyama, T.; Miharada, K.; Sudo, K.; Danjo, I.; Aoki, N.; Nakamura, Y. Establishment of Mouse Embryonic Stem Cell-Derived Erythroid Progenitor Cell Lines Able to Produce Functional Red Blood Cells. PLoS ONE 2008, 3, e1544. [Google Scholar] [CrossRef] [PubMed]
- Ghirlando, R.; Felsenfeld, G. CTCF: Making the Right Connections. Genes Dev. 2016, 30, 881–891. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Wang, X. Roles of Cohesin in Chromosome Architecture and Gene Expression. Semin. Cell Dev. Biol. 2019, 90, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, T.; O’Geen, H.; Keles, S.; Blahnik, K.; Linnemann, A.K.; Kang, Y.A.; Choi, K.; Farnham, P.J.; Bresnick, E.H. Discovering Hematopoietic Mechanisms through Genome-Wide Analysis of GATA Factor Chromatin Occupancy. Mol. Cell 2009, 36, 667–681. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Riva, L.; Xie, H.; Schindler, Y.; Moran, T.B.; Cheng, Y.; Yu, D.; Hardison, R.; Weiss, M.J.; Orkin, S.H.; et al. Insights into GATA-1-Mediated Gene Activation Versus Repression Via Genome-Wide Chromatin Occupancy Analysis. Mol. Cell 2009, 36, 682–695. [Google Scholar] [CrossRef]
- Soler, E.; Andrieu-Soler, C.; de Boer, E.; Bryne, J.C.; Thongjuea, S.; Stadhouders, R.; Palstra, R.J.; Stevens, M.; Kockx, C.; van Ijcken, W.; et al. The Genome-Wide Dynamics of the Binding of Ldb1 Complexes during Erythroid Differentiation. Genes Dev. 2010, 24, 277–289. [Google Scholar] [CrossRef]
- Yue, F.; Cheng, Y.; Breschi, A.; Vierstra, J.; Wu, W.; Ryba, T.; Sandstrom, R.; Ma, Z.; Davis, C.; Pope, B.D.; et al. A Comparative Encyclopedia of DNA Elements in the Mouse Genome. Nature 2014, 515, 355–364. [Google Scholar] [CrossRef]
- Robson, M.I.; Ringel, A.R.; Mundlos, S. Regulatory Landscaping: How Enhancer-Promoter Communication is Sculpted in 3D. Mol. Cell 2019, 74, 1110–1122. [Google Scholar] [CrossRef]
- Zheng, H.; Xie, W. The Role of 3D Genome Organization in Development and Cell Differentiation. Nat. Rev. Mol. Cell Biol. 2019, 20, 535–550. [Google Scholar] [CrossRef] [PubMed]
- Nora, E.P.; Lajoie, B.R.; Schulz, E.G.; Giorgetti, L.; Okamoto, I.; Servant, N.; Piolot, T.; van Berkum, N.L.; Meisig, J.; Sedat, J.; et al. Spatial Partitioning of the Regulatory Landscape of the X-Inactivation Centre. Nature 2012, 485, 381–385. [Google Scholar] [CrossRef] [PubMed]
- Dixon, J.R.; Selvaraj, S.; Yue, F.; Kim, A.; Li, Y.; Shen, Y.; Hu, M.; Liu, J.S.; Ren, B. Topological Domains in Mammalian Genomes Identified by Analysis of Chromatin Interactions. Nature 2012, 485, 376–380. [Google Scholar] [CrossRef] [PubMed]
- Phillips-Cremins, J.E.; Sauria, M.E.; Sanyal, A.; Gerasimova, T.I.; Lajoie, B.R.; Bell, J.S.; Ong, C.T.; Hookway, T.A.; Guo, C.; Sun, Y.; et al. Architectural Protein Subclasses Shape 3D Organization of Genomes during Lineage Commitment. Cell 2013, 153, 1281–1295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Catarino, R.R.; Stark, A. Assessing Sufficiency and Necessity of Enhancer Activities for Gene Expression and the Mechanisms of Transcription Activation. Genes Dev. 2018, 32, 202–223. [Google Scholar] [CrossRef] [PubMed]
- Dore, L.C.; Chlon, T.M.; Brown, C.D.; White, K.P.; Crispino, J.D. Chromatin Occupancy Analysis Reveals Genome-Wide GATA Factor Switching during Hematopoiesis. Blood 2012, 119, 3724–3733. [Google Scholar] [CrossRef]
- Papadopoulos, G.L.; Karkoulia, E.; Tsamardinos, I.; Porcher, C.; Ragoussis, J.; Bungert, J.; Strouboulis, J. GATA-1 Genome-Wide Occupancy Associates with Distinct Epigenetic Profiles in Mouse Fetal Liver Erythropoiesis. Nucleic Acids Res. 2013, 41, 4938–4948. [Google Scholar] [CrossRef] [PubMed]
- Vakoc, C.R.; Letting, D.L.; Gheldof, N.; Sawado, T.; Bender, M.A.; Groudine, M.; Weiss, M.J.; Dekker, J.; Blobel, G.A. Proximity among Distant Regulatory Elements at the Beta-Globin Locus Requires GATA-1 and FOG-1. Mol. Cell 2005, 17, 453–462. [Google Scholar] [CrossRef]
- Lee, J.; Krivega, I.; Dale, R.K.; Dean, A. The LDB1 Complex Co-Opts CTCF for Erythroid Lineage-Specific Long-Range Enhancer Interactions. Cell Rep. 2017, 19, 2490–2502. [Google Scholar] [CrossRef]
- Kim, S.I.; Bultman, S.J.; Kiefer, C.M.; Dean, A.; Bresnick, E.H. BRG1 Requirement for Long-Range Interaction of a Locus Control Region with a Downstream Promoter. Proc. Natl. Acad. Sci. USA 2009, 106, 2259–2264. [Google Scholar] [CrossRef]
- Yun, W.J.; Kim, Y.W.; Kang, Y.; Lee, J.; Dean, A.; Kim, A. The Hematopoietic Regulator TAL1 is Required for Chromatin Looping between the Beta-Globin LCR and Human Gamma-Globin Genes to Activate Transcription. Nucleic Acids Res. 2014, 42, 4283–4293. [Google Scholar] [CrossRef] [PubMed]
- Takano, H.; Nakazawa, S.; Okuno, Y.; Shirata, N.; Tsuchiya, S.; Kainoh, T.; Takamatsu, S.; Furuta, K.; Taketomi, Y.; Naito, Y.; et al. Establishment of the Culture Model System that Reflects the Process of Terminal Differentiation of Connective Tissue-Type Mast Cells. FEBS Lett. 2008, 582, 1444–1450. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, L.; Tsukamoto, S.; Suzuki, M.; Yamamoto-Mukai, H.; Yamamoto, M.; Philipsen, S.; Ohneda, K. Ablation of Gata1 in Adult Mice Results in Aplastic Crisis, Revealing its Essential Role in Steady-State and Stress Erythropoiesis. Blood 2008, 111, 4375–4385. [Google Scholar] [CrossRef] [PubMed]
- Charles, M.A.; Saunders, T.L.; Wood, W.M.; Owens, K.; Parlow, A.F.; Camper, S.A.; Ridgway, E.C.; Gordon, D.F. Pituitary-Specific Gata2 Knockout: Effects on Gonadotrope and Thyrotrope Function. Mol. Endocrinol. 2006, 20, 1366–1377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mizushima, S.; Nagata, S. PEF-BOS, a Powerful Mammalian Expression Vector. Nucleic Acids Res. 1990, 18, 5322. [Google Scholar] [CrossRef]
- Shimizu, R.; Takahashi, S.; Ohneda, K.; Engel, J.D.; Yamamoto, M. In Vivo Requirements for GATA-1 Functional Domains during Primitive and Definitive Erythropoiesis. EMBO J. 2001, 20, 5250–5260. [Google Scholar] [CrossRef] [PubMed]
- Ishijima, Y.; Ohmori, S.; Uenishi, A.; Ohneda, K. GATA Transcription Factors are Involved in IgE-Dependent Mast Cell Degranulation by Enhancing the Expression of Phospholipase C-gamma1. Genes Cells 2012, 17, 285–301. [Google Scholar] [CrossRef]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome Engineering using the CRISPR-Cas9 System. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef]





© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ohneda, K.; Ohmori, S.; Yamamoto, M. Mouse Tryptase Gene Expression is Coordinately Regulated by GATA1 and GATA2 in Bone Marrow-Derived Mast Cells. Int. J. Mol. Sci. 2019, 20, 4603. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20184603
Ohneda K, Ohmori S, Yamamoto M. Mouse Tryptase Gene Expression is Coordinately Regulated by GATA1 and GATA2 in Bone Marrow-Derived Mast Cells. International Journal of Molecular Sciences. 2019; 20(18):4603. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20184603
Chicago/Turabian StyleOhneda, Kinuko, Shin’ya Ohmori, and Masayuki Yamamoto. 2019. "Mouse Tryptase Gene Expression is Coordinately Regulated by GATA1 and GATA2 in Bone Marrow-Derived Mast Cells" International Journal of Molecular Sciences 20, no. 18: 4603. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20184603




