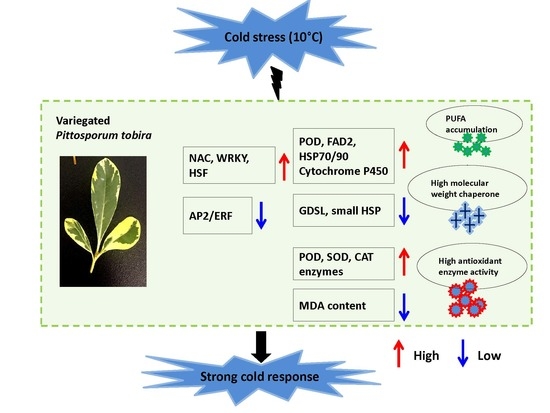
Protective Role of Leaf Variegation in Pittosporum tobira under Low Temperature: Insights into the Physio-Biochemical and Molecular Mechanisms
Abstract
:1. Introduction
2. Results
2.1. Characteristics of Variegated Leaves in Pittosporum Tobira
2.2. Effect of Temperature Decrease on ROS-Scavenging Enzyme Activities in P. tobira Cultivars
2.3. Transcriptome Sequencing in Leaves of “Variegatum” and “Green Pittosporum” and Functional Annotation of Unigenes
2.4. Differential Gene Expression Analysis between “Variegatum” and “Green Pittosporum” under Cold Condition
2.5. Major Transcription Factors Differentially Regulated between “Variegatum” and “Green Pittosporum” under Cold Conditions
2.6. DEGs Related to the Biosynthesis of Unsaturated Fatty Acids and Fatty Acids Metabolism
2.7. Disturbance of Protein Processing in Endoplasmic Reticulum under Cold Conditions
2.8. DEGs in the Phenylalanine Metabolism
3. Discussion
3.1. Characteristics of Leaf Variegation in P. tobira
3.2. Protective Role of Leaf Variegation in P. tobira under Cold Condition
4. Materials and Methods
4.1. Plant Materials
4.2. Transmission Electron Microscopy (TEM)
4.3. Measurement of Physio-Biochemical Parameters
4.4. RNA Extraction, cDNA Library Construction, and Transcriptome Sequencing
4.5. De novo Assembly, Functional Annotation, Classification and Metabolic Pathway Analysis
4.6. Differential Expression and Enrichment Analysis
4.7. Validation of Gene Expression Using Quantitative Real Time-PCR
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| CAT | Catalase |
| DEG | Differentially expressed gene |
| FAD2 | Fatty acid desaturase 2 |
| FPKM | Fragments per kilobase of exon per million fragments mapped |
| GO | Gene ontology |
| HSF | Heat shock factor |
| HSP | Heat shock protein |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| MDA | Malonaldehyde |
| POD | Peroxidase |
| PUFA | Poly-unsaturated fatty acids |
| ROS | Reactive oxygen species |
| SOD | Superoxide dismutase |
| TF | Transcription factor |
References
- Aluru, M.R.; Bae, H.; Wu, D.; Rodermel, S.R. The Arabidopsis immutans mutation affects plastid differentiation and the morphogenesis of white and green sectors in variegated plants. Plant Physiol. 2001, 127, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Kato, Y.; Kouso, T.; Sakamoto, W. Variegated tobacco leaves generated by chloroplast FtsH suppression: Implication of FtsH function in the maintenance of thylakoid membranes. Plant Cell Physiol. 2012, 53, 391–404. [Google Scholar] [CrossRef] [PubMed]
- Shih, T.-H.; Lin, S.-H.; Huang, M.-Y.; Huang, W.-D.; Yang, C.-M. Transcriptome profile of the variegated Ficus microcarpa c.v. milkys fig leaf. Int. J. Mol. Sci. 2019, 20, 1338. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.C.; Wu, Y.J.; Sheue, C.R.; Liao, P.C.; Chen, Y.H.; Li, S.J.; Liu, J.W.; Chang, H.T.; Liu, W.L.; Ko, Y.Z.; et al. Molecular basis underlying leaf variegation of a moth orchid mutant (Phalaenopsis aphrodite subsp. formosana). Front. Plant Sci. 2017, 8, 1333. [Google Scholar] [CrossRef] [PubMed]
- Hara, N. Study of the variegated leaves with special reference to those caused by air spaces. Jpn. J. Bot. 1957, 16, 86–101. [Google Scholar]
- Sheue, C.R.; Pao, S.H.; Chien, L.F.; Chesson, P.; Peng, C.I. Natural foliar variegation without costs? The case of Begonia. Ann. Bot. 2012, 109, 1065–1074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsukaya, H.; Okada, H.; Mohamed, M. A novel feature of structural variegation in leaves of the tropical plant Schismatoglottis calyptrata. J. Plant Res. 2004, 117, 477–480. [Google Scholar] [CrossRef]
- Tilney-Bassett, R.A.E. Genetics of variegated plants. In Genetics and Biogenesis of Mitochondria and Chloroplasts; Birky, C.W., Perlman, P.S., Byers, T.J., Eds.; Ohio State University Press: Columbus, OH, USA, 1975; pp. 268–308. [Google Scholar]
- Beardsell, D.; Norden, U. Ficus rubiginosa ‘variegata’, a chlorophyll-deficient chimera with mosaic patterns created by cell divisions from the outer meristematic layer. Ann. Bot. 2004, 94, 51–58. [Google Scholar] [CrossRef]
- Yu, F.; Fu, A.; Aluru, M.; Park, S.; Xu, Y.; Liu, H.; Liu, X.; Foudree, A.; Nambogga, M.; Rodermel, S. Variegation mutants and mechanisms of chloroplast biogenesis. Plant Cell Environ. 2007, 30, 350–365. [Google Scholar] [CrossRef]
- Li, X.; Kanakala, S.; He, Y.; Zhong, X.; Yu, S.; Li, R.; Sun, L.; Ma, J. Physiological characterization and comparative transcriptome analysis of white and green leaves of Ananas comosus var. bracteatus. PLoS ONE 2017, 12, e0169838. [Google Scholar] [CrossRef]
- Lysenko, V. Fluorescence kinetic parameters and cyclic electron transport in guard cell chloroplasts of chlorophyll-deficient leaf tissues from variegated weeping fig (Ficus benjamina L.). Planta 2012, 235, 1023–1033. [Google Scholar] [CrossRef] [PubMed]
- Aluru, M.R.; Zola, J.; Foudree, A.; Rodermel, S.R. Chloroplast photooxidation-induced transcriptome reprogramming in Arabidopsis immutans white leaf sectors. Plant Physiol. 2009, 150, 904–923. [Google Scholar] [CrossRef] [PubMed]
- Miura, E.; Kato, Y.; Sakamoto, W. Comparative transcriptome analysis of green/white variegated sectors in Arabidopsis yellow variegated2: Responses to oxidative and other stresses in white sectors. J. Exp. Bot. 2010, 61, 2433–2445. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Chen, X.; Xu, B.; Li, Y.; Ma, Y.; Wang, G. Phenotype and transcriptome analysis reveals chloroplast development and pigment biosynthesis together influenced the leaf color formation in mutants of Anthurium and raeanum ‘Sonate’. Front. Plant Sci. 2015, 6, 139. [Google Scholar] [CrossRef] [PubMed]
- Lev-Yadun, S.; Dafni, A.; Flaishman, M.A.; Inbar, M.; Izhaki, I.; Katzir, G.; Ne’eman, G. Plant coloration undermines herbivorous insect camouflage. BioEssays 2004, 26, 1126–1130. [Google Scholar] [CrossRef] [PubMed]
- Lev-Yadun, S. Defensive (anti-herbivory) coloration in land plants. In Anti- Herbivory Plant Coloration and Morphology; Springer: Zug, Switzerland, 2016. [Google Scholar]
- Lev-Yadun, S. Local loss of the zebra-like coloration supports the aposematic and other visual defense hypotheses in Silybum marianum. Isr. J. Plant Sci. 2017, 64, 170–178. [Google Scholar] [CrossRef]
- Niu, Y.; Sun, H.; Stevens, M. Plant camouflage: Ecology, evolution, and implications. Trends Ecol. Evol. 2018, 33, 608–618. [Google Scholar] [CrossRef]
- Fooshee, W.C.; Henny, R.J. Chlorophyll levels and anatomy of variegated and non variegated areas of Aglaonema nitidum leaves. Proc. Fla. State Hortic. Soc. 1990, 103, 170–172. [Google Scholar]
- Roelfsema, M.; Konrad, K.R.; Marten, H.; Psaras, G.K.; Hartung, W.; Hedrich, R. Guard cells in albino leaf patches do not respond to photosynthetically active radiation, but are sensitive to blue light, CO2 and abscisic acid. Plant Cell Environ. 2006, 29, 1595–1605. [Google Scholar] [CrossRef]
- Shelef, O.; Summerfield, L.; Lev-Yadun, S.; Villamarin-Cortez, S.; Sadeh, R.; Herrmann, I.; Rachmilevitch, S. Thermal benefits from white variegation of Silybum marianum Leaves. Front. Plant Sci. 2019, 10, 688. [Google Scholar] [CrossRef]
- Smith, A.P. Ecology of a leaf color polymorphism in a tropical forest species: Habitat segregation and herbivory. Oecologia 1986, 69, 283–287. [Google Scholar] [CrossRef] [PubMed]
- Soltau, U.; Dötterl, S.; Liede-Schumann, S. Leaf variegation in Caladium steudneriifolium (Araceae): A case of mimicry? Evol. Ecol. 2009, 23, 503–512. [Google Scholar] [CrossRef]
- Mwafongo, E.; Vollsnesb, A.V.; Bjorå, C.S.; Nordal, I.; Eriksen, A.B. Leaf mottling/variegation and shape in the Ledebouria revoluta complex—Development, stability and putative function. Flora 2017, 236, 33–43. [Google Scholar] [CrossRef]
- Dib, R.A.E.; Eskander, J.; Mohamed, M.A.; Mohammed, N.M. Two new triterpenoid estersaponins and biological activities of Pittosporum tobira, ‘variegata’ (Thunb.) WT. Aiton leaves. Fitoterapia 2015, 106, 272–279. [Google Scholar] [CrossRef]
- Burt, S. Essential oils: Their antibacterial properties and potential applications in foods-a review. Int. J. Food Microbiol. 2004, 94, 223–253. [Google Scholar] [CrossRef]
- Eunsil, K.O.; Choi, M.R.; Choi, K.M.; Cha, J.D. The effect of Pittosporum tobira against anti-Helicobacter pylori and anti-oxidant activity. IJVR 2014, 8, 4. [Google Scholar]
- Jiang, H.; Chen, Y.; Li, M.; Xu, X.; Wu, G. Overexpression of SGR results in oxidative stress and lesion-mimic cell death in rice seedlings. J. Integr. Plant Biol. 2011, 53, 375–387. [Google Scholar] [CrossRef]
- Han, S.H.; Sakuraba, Y.; Koh, H.J.; Paek, N.C. Leaf variegation in the rice zebra2 mutant is caused by photoperiodic accumulation of tetra-Cis-lycopene and singlet oxygen. Mol. Cells 2012, 33, 87–97. [Google Scholar] [CrossRef]
- Sheoran, S.; Thakur, V.; Narwal, S.; Turan, R.; Mamrutha, H.M.; Singh, V.; Tiwari, V.; Sharma, I. Differential activity and expression profile of antioxidant enzymes and physiological changes in wheat (Triticum aestivum L.) under drought. Appl. Biochem. Biotechnol. 2015, 177, 1282–1298. [Google Scholar] [CrossRef]
- Esteban, R.; Fernandez-Marin, B.; Becerril, J.M.; Garcia-Plazaola, J.I. Photoprotective implications of leaf variegation in E. denscanis L. and P. officinalis L. J. Plant Physiol. 2007, 165, 1255–1263. [Google Scholar] [CrossRef]
- Sui, N.; Li, M.; Zhao, S.J.; Li, F.; Liang, H.; Meng, Q.W. Overexpression of glycerol-3-phosphate acyltransferase gene improves chilling tolerance in tomato. Planta 2007, 226, 1097–1108. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-H.; Kwon, C.-T.; Song, G.; Koh, H.-G.; An, G.; Paek, N.-C. The rice zebra3 (z3) mutation disrupts citrate distribution and produces transverse dark-green/green variegation in mature leaves. Rice 2018, 11, 1. [Google Scholar] [CrossRef] [PubMed]
- Wetzel, C.M.; Jiang, C.Z.; Meehan, L.J.; Voytas, D.F.; Rodermel, S.R. Nuclear-organelle interactions: The immutans variegation mutant of Arabidopsis is plastid autonomous and impaired in carotenoid biosynthesis. Plant J. 1994, 6, 161–175. [Google Scholar] [CrossRef] [PubMed]
- Miura, E.; Kato, Y.; Matsushima, R.; Albrecht, V.; Laalami, S.; Sakamoto, W. The balance between protein synthesis and degradation in chloroplasts determines leaf variegation in Arabidopsis yellow variegated mutants. Plant Cell 2007, 19, 1313–1328. [Google Scholar] [CrossRef] [PubMed]
- Bae, C.H.; Abe, T.; Matsuyama, T.; Fukunishi, N.; Nagata, N.; Nakano, T.; Kaneko, Y.; Miyoshi, K.; Matsushima, H.; Yoshida, S. Regulation of chloroplast gene expression is affected in ali, a novel tobacco albino mutant. Ann. Bot. 2001, 88, 545–553. [Google Scholar] [CrossRef]
- Aluru, M.R.; Yu, F.; Fu, A.; Rodermel, S. Arabidopsis variegation mutants: New insights into chloroplast biogenesis. J. Exp. Bot. 2006, 57, 1871–1881. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.-H.; Hung, C.-Y.; Qiu, J.; Chen, J.; Kittur, F.S.; Oldham, C.E.; Henny, R.J.; Burkey, K.O.; Fan, L.; Xie, J. Accumulation of high OPDA level correlates with reduced ROS and elevated GSH benefiting white cell survival in variegated leaves. Sci. Rep. 2017, 7, 44158. [Google Scholar] [CrossRef]
- Klančnik, K.; Levpušček, M.; Gaberščik, A. Variegation and red abaxial epidermis define the leaf optical properties of Cyclamen purpurascens. Flora 2016, 224, 87–95. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhang, M.; Ding, Y.; Zhou, P.; Fang, Y. Composition of photosynthetic pigments and photosynthetic characteristics in green and yellow sectors of the variegated Aucuba japonica Variegata leaves. Flora 2018, 240, 25–33. [Google Scholar] [CrossRef]
- Borek, M.; Baczek-Kwinta, R.; Rapacz, M. Photosynthetic activity of variegated leaves of Coleus x hybridus hort. cultivars characterised by chlorophyll fluorescence techniques. Photosynthetica 2016, 54, 331–339. [Google Scholar] [CrossRef]
- Hashimoto, H.; Uragami, C.; Cogdell, R.J. Carotenoids and Photosynthesis. In Carotenoids in Nature; Subcellular Biochemistry; Stange, C., Ed.; Springer: Cham, Switzerland, 2016; Volume 79. [Google Scholar] [CrossRef]
- Farooq, M.; Hussain, M.; Wahid, A.; Siddique, K.H.M. Drought stress in plants: An overview. In Plant Responses to Drought Stress: From Morphological to Molecular Features; Aroca, R., Ed.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 1–33. [Google Scholar]
- Mikami, K.; Murata, N. Membrane fluidity and the perception of environmental signals in cyanobacteria and plants. Prog. Lipid Res. 2003, 42, 527–543. [Google Scholar] [CrossRef]
- Dakhma, W.S.; Zarrouk, M.; Cherif, A. Effects of drought stress on lipids in rape leaves. Phytochemistry 1995, 40, 1383–1386. [Google Scholar] [CrossRef]
- Olsson, M. Alteration in lipid-composition, lipid-peroxidation and antioxidative protection during senescence in drought stressed plants of Pisum sativum. Plant Physiol. Biochem. 1995, 33, 547–553. [Google Scholar]
- Matos, M.C.; Campos, P.S.; Ramalho, J.C.; Medeira, M.C.; Maia, M.I.; Semedo, J.M.; Marques, N.M.; Matos, A. Photosynthetic activity and cellular integrity of the Andean legume Pachyrhizus ahipa (Wedd.) Parodi under heat and water stress. Photosynthetica 2002, 40, 493–501. [Google Scholar] [CrossRef]
- Sui, N.; Li, M.; Shu, D.F.; Zhao, S.J.; Meng, Q.W. Antisense mediated depletion of tomato chloroplast glycerol-3- phosphate acyltransferase affects male fertility and increases thermal tolerance. Physiol. Plant. 2007, 130, 301–314. [Google Scholar] [CrossRef]
- Sui, N.; Li, M.; Li, K.; Song, J.; Wang, B.-S. Increase in unsaturated fatty acids in membrane lipids of Suaeda salsa L. enhances protection of photosystem II under high salinity. Photosynthetica 2010, 48, 623–629. [Google Scholar] [CrossRef]
- Liu, X.-Y.; Li, B.; Yang, J.-H.; Sui, N.; Yang, X.-M.; Meng, Q.-W. Overexpression of tomato chloroplast omega-3 fatty acid desaturase gene alleviates the photoinhibition of photosystems 2 and 1 under chilling stress. Photosynthetica 2008, 46, 185–192. [Google Scholar] [CrossRef]
- Upchurch, R.G. Fatty acid unsaturation, mobilization, and regulation in the response of plants to stress. Biotechnol. Lett. 2008, 30, 967–977. [Google Scholar] [CrossRef] [PubMed]
- Dar, A.; Choudhury, A.R.; Kancharla, P.K.; Arumugam, N. The FAD2 gene in plants: Occurrence, regulation, and role. Front. Plant Sci. 2017, 8, 1789. [Google Scholar] [CrossRef] [PubMed]
- Los, D.A.; Murata, N. Structure and expression of fatty acid desaturases. Biochem. Biophys. Acta 1998, 1394, 3–15. [Google Scholar] [CrossRef]
- Routaboul, J.M.; Fischer, S.F.; Browse, J. Trienoic fatty acids are required to maintain chloroplast function at low temperatures. Plant Physiol. 2000, 124, 1697–1705. [Google Scholar] [CrossRef] [PubMed]
- Vega, S.E.; del Rio, A.H.; Bamberg, J.B.; Palta, J.P. Evidence for the up-regulation of stearoyl-ACP (D9) desaturase gene expression during cold acclimation. Am. J. Potato Res. 2004, 81, 125–135. [Google Scholar] [CrossRef]
- Kargiotidou, A.; Deli, D.; Galanopoulou, D.; Tsaftaris, A.; Farmaki, T. Low temperature and light regulate delta 12 fatty acid desaturases (FAD2) at a transcriptional level in cotton (Gossypium hirsutum). J. Exp. Bot. 2008, 59, 2043–2056. [Google Scholar] [CrossRef] [PubMed]
- Kreps, J.A.; Wu, Y.; Chang, H.S.; Zhu, T.; Wang, X.; Harper, J.F. Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol. 2002, 130, 129–2141. [Google Scholar] [CrossRef] [PubMed]
- Matteucci, M.; D’Angeli, S.; Errico, S.; Lamanna, R.; Perrotta, G.; Altamura, M.M. Cold affects the transcription of fatty acid desaturases and oil quality in the fruit of Olea europaea L. genotypes with different cold hardiness. J. Exp. Bot. 2011, 62, 3403–3420. [Google Scholar] [CrossRef] [PubMed]
- Los, D.A.; Ray, M.K.; Murata, N. Differences in the control of the temperature-dependent expression of four genes for desaturases in Synechocystis sp. PCC 6803. Mol. Microbiol. 1997, 25, 1167–1175. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.M.; Lai, C.P.; Chen LF, O.; Chan, M.T.; Shaw, J.F. Arabidopsis SFAR4 is a novel GDSL-type esterase involved in fatty acid degradation and glucose tolerance. Botani Stud. 2015, 56, 33. [Google Scholar] [CrossRef]
- Xu, H.M.; Kong, X.D.; Chen, F.; Huang, J.X.; Lou, X.Y.; Zhao, J.Y. Transcriptome analysis of Brassica napus pod using RNA-Seq and identification of lipid-related candidate genes. BMC Genom. 2015, 16, 858. [Google Scholar] [CrossRef]
- Duan, S.; Jin, C.; Li, D.; Gao, C.; Qi, S.; Liu, K.; Hai, J.; Ma, H.; Chen, M. MYB76 inhibits seed fatty acid accumulation in Arabidopsis. Front. Plant Sci. 2017, 8, 226. [Google Scholar] [CrossRef]
- Zhang, Y.; Bai, B.; Lee, M.; Alfiko, Y.; Suwanto, A.; Yue, G.H. Cloning and characterization of EgGDSL, a gene associated with oil content in oil palm. Sci. Rep. 2018, 8, 11406. [Google Scholar] [CrossRef]
- Al-Whaibi, M.H. Plant heat-shock proteins: A mini review. J. King Saud Univ. Sci. 2011, 23, 139–150. [Google Scholar] [CrossRef] [Green Version]
- Vierling, E. The Roles of Heat Shock Proteins in Plants. Ann. Rev. Plant Physiol. Plant Mol. Biol. 1991, 42, 579–620. [Google Scholar] [CrossRef]
- Krishna, P.; Sacco, M.; Cherutti, J.F.; Hill, S. Cold induced accumulation of Hsp90 transcripts in Brassica napus. Plant Physiol. 1995, 107, 915–923. [Google Scholar] [CrossRef] [PubMed]
- Pareek, A.; Singla, S.L.; Grover, A. Plant Hsp90 family with special reference to rice. J. Biosci. 1998, 23, 361–367. [Google Scholar] [CrossRef]
- Kilian, J.; Whitehead, D.; Horak, J.; Wanke, D.; Weinl, S.; Batistic, O.; Bauer, E.B.; D’Angelo; Kudla, J.; Harter, K. The AtGenExpress global stress expression data set: Protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 2007, 50, 347–363. [Google Scholar] [CrossRef]
- Zhang, J.-H.; Wang, L.-J.; Pan, Q.-H.; Wang, Y.-Z.; Zhan, J.-C.; Huang, W.-D. Accumulation and subcellular localization of heat shock proteins in young grape leaves during cross-adaptation to temperature stresses. Sci. Hortic. 2008, 117, 231–240. [Google Scholar] [CrossRef]
- Baniwal, S.K.; Bharti, K.; Chan, K.Y.; Fauth, M.; Ganguli, A.; Kotak, S.; Mishra, S.K.; Nover, L.; Port, M.; Scharf, K.; et al. Heat stress response in plants: A complex game with chaperones and more than 20 heat stress transcription factors. J. Biosci. 2004, 29, 471–487. [Google Scholar] [CrossRef]
- Hu, W.; Hu, G.; Han, B. Genome-wide survey and expression profiling of heat shock proteins and heat shock factors revealed overlapped and stress specific response under abiotic stresses in rice. Plant Sci. 2009, 176, 583–590. [Google Scholar] [CrossRef]
- Gao, C.; Wang, Y.; Liu, G.; Wang, C.; Jiang, J.; Yang, C. Cloning of ten peroxidase (POD) genes from Tamarix hispida and characterization of their responses to abiotic stress. Plant Mol. Biol. Report. 2010, 28, 77. [Google Scholar] [CrossRef]
- Kim, S.-H.; Choi, H.-S.; Cho, Y.-C.; Kim, S.-R. Cold-responsive regulation of a flower-preferential class III peroxidase gene, OsPOX1, in rice (Oryza sativa L.). J. Plant Biol. 2012, 55, 123–131. [Google Scholar] [CrossRef]
- Wang, J.-E.; Liu, K.-K.; Li, D.-W.; Zhang, Y.-L.; Zhao, Q.; He, Y.-M.; Gong, Z.-H. A Novel Peroxidase CanPOD Gene of Pepper Is Involved in Defense Responses to Phytophtora capsici Infection as well as Abiotic Stress Tolerance. Int. J. Mol. Sci. 2013, 14, 3158–3177. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Cheng, C.; Chen, F.; Ni, S.; Lin, Y.; Lai, Z. High-throughput sequencing of small RNAs revealed the diversified cold-responsive pathways during cold stress in the wild banana (Musa itinerans). BMC Plant Biol. 2018, 18, 308. [Google Scholar] [CrossRef] [PubMed]
- Wellburn, A.R. The spectral determination of chlorophylls a and b, as well as total carotenoids, using various solvents with spectrophotometers of different resolution. J. Plant Physiol. 1994, 144, 307–313. [Google Scholar] [CrossRef]
- Zhuang, H.; Lou, Q.; Liu, H.; Han, H.; Wang, Q.; Tang, Z.; Ma, Y.; Wang, H. Differential Regulation of Anthocyanins in Green and Purple Turnips Revealed by Combined De Novo Transcriptome and Metabolome Analysis. Int. J. Mol. Sci. 2019, 20, 4387. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full length transcriptome assembly from RNA Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S.; Kawashima, S.; Okuno, Y.; Hattori, M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004, 32, D277–D280. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Tatusov, R.L.; Galperin, M.Y.; Natale, D.A. The COG database: A tool for genome scale analysis of protein functions and evolution. Nucleic Acids Res. 2000, 28, 33–36. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2013, 42, D222–D230. [Google Scholar] [CrossRef]
- Apweiler, R.; Bairoch, A.; Wu, C.H.; Barker, W.C.; Boeckmann, B.; Ferro, S.; Gasteiger, E.; Huang, H.; Lopez, R.; Magrane, M.; et al. UniProt: The Universal Protein knowledgebase. Nucleic Acids Res. 2004, 32, D115–D119. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Szklarczyk, D.; Forslund, K.; Cook, H.; Heller, D.; Walter, M.C.; Rattei, T.; Mende, D.R.; Sunagawa, S.; Kuhn, M.; et al. eggNOG 4.5: A hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Research 2015, 44, D286–D293. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.Y.; Li, J.Q.; Wu, S.F.; Zhu, Y.; Chen, Y.W.; He, F.C. Integrated nr Database in Protein Annotation System and Its Localization. Comput. Eng. 2006, 32, 71–74. [Google Scholar]
- Koonin, E.V.; Fedorova, N.D.; Jackson, J.D.; Jacobs, A.R.; Krylov, D.M.; Makarova, K.S.; Mazumder, R.; Mekhedov, S.L.; Nikolskaya, A.N.; Rao, B.S.; et al. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol. 2004, 5, R7. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI BLAST: A New Generation of Protein Database Search Programs. Nucleic Acids Res. Ital. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef] [PubMed]
- Eddy, S.R. Profile hidden Markov models. Bioinform. Ital. 1998, 14, 755–763. [Google Scholar]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Colin, N.D. RSEM: Accurate transcript quantification from RNA Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Anders, S.; McCarthy, D.J.; Chen, Y.; Okoniewski, M.; Smyth, G.K.; Huber, W.; Robinson, M.D. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat. Protoc. 2013, 8, 1765–1786. [Google Scholar] [CrossRef]
- Alexa, A.; Rahnenfuhrer, J. topGO: Enrichment Analysis for Gene Ontology; R Package Version 2.18.0; 2010; Volume 2, p. 2010. [Google Scholar]
- Dossa, K.; Li, D.J.; Yu, J.; Wang, L.; Zhang, Y.; You, J.; Zhou, R.; Mmadi, M.A.; Li, A.; Fonceka, D.; et al. The genetic basis of drought tolerance in the high oil crop Sesamum indicum. Plant Biotechnol. J. 2019, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Lalitha, S. Primer premier 5. Biotechnol. Softw Internet Rep. 2000, 1, 270–272. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]








| Cultivar | Library-ID | Read Number | Base Number | GC (%) | % ≥ Q30 |
|---|---|---|---|---|---|
| “Green Pittosporum” | T01 | 22,115,602 | 6,612,391,012 | 45.27 | 92.78 |
| “Green Pittosporum” | T02 | 23,965,370 | 7,164,119,518 | 45.01 | 92.97 |
| “Green Pittosporum” | T03 | 22,120,563 | 6,612,379,716 | 45.25 | 92.96 |
| “Variegatum” | T04 | 23,413,612 | 6,996,233,542 | 44.79 | 93.60 |
| “Variegatum” | T05 | 22,179,179 | 6,627,936,224 | 44.64 | 93.36 |
| “Variegatum” | T06 | 22,956,424 | 6,863,508,574 | 45.19 | 92.86 |
| Length Range | Transcript | Unigene |
|---|---|---|
| 200–300 | 53,860(25.64%) | 46,279(41.00%) |
| 300–500 | 38,333(18.25%) | 27,916(24.73%) |
| 500–1000 | 38,190(18.18%) | 20,115(17.82%) |
| 1000–2000 | 39,154(18.64%) | 11,929(10.57%) |
| 2000 | 40,551(19.30%) | 6,636(5.88%) |
| Total number | 210,088 | 112,875 |
| Total length (bp) | 241,105,749 | 72,533,944 |
| N50 length (bp) | 2,137 | 1,017 |
| Mean length (bp) | 1147.64 | 642.60 |
| #Anno_Database | Annotated_Number | 300 <= length < 1000 | Length >= 1000 |
|---|---|---|---|
| COG_Annotation | 17,065 | 6,007 | 6,317 |
| GO_Annotation | 28,283 | 10,378 | 9,062 |
| KEGG_Annotation | 19,595 | 7,857 | 6,277 |
| KOG_Annotation | 30,496 | 11,470 | 10,018 |
| Pfam_Annotation | 33,826 | 12,283 | 13,569 |
| Swissprot_Annotation | 28,074 | 10,455 | 11,043 |
| eggNOG_Annotation | 48,410 | 18,186 | 15,546 |
| nr_Annotation | 47,309 | 17,751 | 15,596 |
| All_Annotated | 51,718 | 19,526 | 15,881 |
| Cultivar | Library-ID | Clean Reads | Mapped Reads | Mapped Ratio |
|---|---|---|---|---|
| “Green Pittosporum” | T01 | 22,115,602 | 17,518,471 | 79.21% |
| “Green Pittosporum” | T02 | 23,965,370 | 19,005,314 | 79.30% |
| “Green Pittosporum” | T03 | 22,120,563 | 17,599,921 | 79.56% |
| “Variegatum” | T04 | 23,413,612 | 19,153,207 | 81.80% |
| “Variegatum” | T05 | 22,179,179 | 17,804,804 | 80.28% |
| “Variegatum” | T06 | 22,956,424 | 18,290,888 | 79.68% |
| Pathway | KO | Gene ID | Log2 Fold Change | Gene Description |
|---|---|---|---|---|
| Phenylalanine metabolism | ||||
| K00815 | c55523.graph_c0 | 1.565 | Aminotransferase TAT2 | |
| K00430 | c68309.graph_c0 | 1.057 | Peroxidase | |
| K00430 | c74970.graph_c1 | 1.157 | Peroxidase | |
| Sesquiterpenoid and triterpenoid biosynthesis | ||||
| K15472 | c29794.graph_c0 | 1.488 | Premnaspirodiene oxygenase, Cytochrome P450 | |
| K15472 | c43399.graph_c0 | 1.198 | Premnaspirodiene oxygenase, Cytochrome P450 | |
| Biosynthesis of unsaturated fatty acids and fatty acid metabolism | ||||
| K10256 | c45880.graph_c0 | 1.973 | FAD2 | |
| K10256 | c72696.graph_c0 | 1.287 | FAD2 | |
| K10256 | c74068.graph_c0 | 1.493 | FAD2 | |
| K10256 | c74296.graph_c0 | 1.385 | FAD2 | |
| K10256 | c75682.graph_c0 | 1.284 | FAD2 | |
| K10256 | c45880.graph_c0 | 1.973 | FAD2 | |
| K10256 | c72696.graph_c0 | 1.287 | FAD2 | |
| K10256 | c74068.graph_c0 | 1.493 | FAD2 | |
| K10256 | c74296.graph_c0 | 1.385 | FAD2 | |
| K10256 | c75682.graph_c0 | 1.284 | FAD2 | |
| −− | c52980.graph_c1 | −1.179 | GDSL | |
| −− | c52915.graph_c0 | −1.582 | GDSL | |
| −− | c62735.graph_c0 | −2.134 | GDSL | |
| −− | c28930.graph_c0 | −1.998 | GDSL | |
| −− | c79198.graph_c0 | −1.068 | GDSL | |
| −− | c63752.graph_c0 | −1.502 | GDSL | |
| −− | c28455.graph_c0 | −1.267 | GDSL | |
| −− | c71358.graph_c0 | −1.635 | GDSL | |
| −− | c76303.graph_c0 | −1.713 | GDSL | |
| Protein processing in endoplasmic reticulum | ||||
| K13993 | c55937.graph_c0 | −2.839 | 22.7kDa HSP IV | |
| K13993 | c60091.graph_c0 | −1.125 | 15 kDa HSP | |
| K13993 | c60491.graph_c0 | −1.961 | 17.9 kDa HSP II | |
| K13993 | c70233.graph_c1 | −1.292 | 18.1 kDa HSP I | |
| K04079 | c74041.graph_c0 | 1.283 | 90 kDa HSP | |
| K03283 | c75081.graph_c0 | 1.095 | 70 kDa HSP | |
| K09489 | c68688.graph_c0 | 1.266 | 70 kDa HSP | |
| K03283 | c28616.graph_c1 | 1.018 | 70 kDa HSP | |
| K03283 | c69810.graph_c6 | 1.408 | 70 kDa HSP | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Z.; Liu, Z.; Song, H.; Chen, M.; Cheng, S. Protective Role of Leaf Variegation in Pittosporum tobira under Low Temperature: Insights into the Physio-Biochemical and Molecular Mechanisms. Int. J. Mol. Sci. 2019, 20, 4857. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20194857
Zhang Z, Liu Z, Song H, Chen M, Cheng S. Protective Role of Leaf Variegation in Pittosporum tobira under Low Temperature: Insights into the Physio-Biochemical and Molecular Mechanisms. International Journal of Molecular Sciences. 2019; 20(19):4857. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20194857
Chicago/Turabian StyleZhang, Zhilu, Zhonghua Liu, Haina Song, Minghui Chen, and Shiping Cheng. 2019. "Protective Role of Leaf Variegation in Pittosporum tobira under Low Temperature: Insights into the Physio-Biochemical and Molecular Mechanisms" International Journal of Molecular Sciences 20, no. 19: 4857. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20194857