Investigating the Molecular Basis of the Aggregation Propensity of the Pathological D76N Mutant of Beta-2 Microglobulin: Role of the Denatured State
Abstract
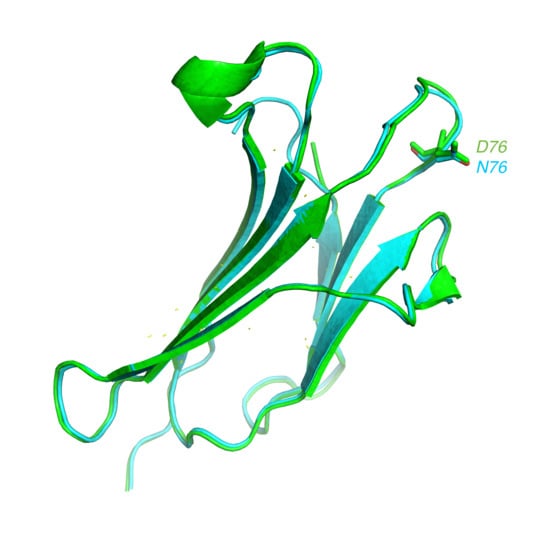
:1. Introduction
2. Results
2.1. Equilibrium Unfolding Experiments
2.2. Aggregation Essays
3. Discussion
4. Materials and Methods
4.1. Protein Expression and Purification
4.2. Equilibrium Experiments
4.3. Aggregation Assays
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Knowles, T.P.; Vendruscolo, M.; Dobson, C.M. The amyloid state and its association with protein misfolding diseases. Nat. Rev. Mol. Cell Biol. 2014, 15, 384–396. [Google Scholar] [CrossRef] [PubMed]
- Westermark, P.; Benson, M.D.; Buxbaum, J.N.; Cohen, A.S.; Frangione, B.; Ikeda, S.; Masters, C.L.; Merlini, G.; Saraiva, M.J.; Sipe, J.D. A primer of amyloid nomenclature. Amyloid 2007, 14, 179–183. [Google Scholar] [CrossRef] [PubMed]
- Chiti, F.; Dobson, C.M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006, 75, 333–366. [Google Scholar] [CrossRef] [PubMed]
- Chiti, F.; Dobson, C.M. Protein Misfolding, Amyloid Formation, and Human Disease: A Summary of Progress Over the Last Decade. Annu. Rev. Biochem. 2017, 86, 27–68. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, A.J.; Knowles, T.P.; Tartaglia, G.G.; Fitzpatrick, A.W.; Devlin, G.L.; Shammas, S.L.; Waudby, C.A.; Mossuto, M.F.; Meehan, S.; Gras, S.L.; et al. Metastability of native proteins and the phenomenon of amyloid formation. J. Am. Chem. Soc. 2011, 133, 14160–14163. [Google Scholar] [CrossRef] [PubMed]
- Stoppini, M.; Bellotti, V. Systemic amyloidosis: Lessons from beta2-microglobulin. J. Biol. Chem. 2015, 290, 9951–9958. [Google Scholar] [CrossRef] [PubMed]
- Gejyo, F.; Yamada, T.; Odani, S.; Nakagawa, Y.; Arakawa, M.; Kunitomo, T.; Kataoka, H.; Suzuki, M.; Hirasawa, Y.; Shirahama, T.; et al. A new form of amyloid protein associated with chronic hemodialysis was identified as beta 2-microglobulin. Biochem. Biophys. Res. Commun. 1985, 129, 701–706. [Google Scholar] [CrossRef]
- Gorevic, P.D.; Casey, T.T.; Stone, W.J.; DiRaimondo, C.R.; Prelli, F.C.; Frangione, B. Beta-2 microglobulin is an amyloidogenic protein in man. J. Clin. Investig. 1985, 76, 2425–2429. [Google Scholar] [CrossRef] [PubMed]
- McParland, V.J.; Kad, N.M.; Kalverda, A.P.; Brown, A.; Kirwin-Jones, P.; Hunter, M.G.; Sunde, M.; Radford, S.E. Partially unfolded states of beta(2)-microglobulin and amyloid formation in vitro. Biochemistry 2000, 39, 8735–8746. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.P.; Jones, S.; Serpell, L.C.; Sunde, M.; Radford, S.E. A systematic investigation into the effect of protein destabilisation on beta 2-microglobulin amyloid formation. J. Mol. Biol. 2003, 330, 943–954. [Google Scholar] [CrossRef]
- Ami, D.; Ricagno, S.; Bolognesi, M.; Bellotti, V.; Doglia, S.M.; Natalello, A. Structure, stability, and aggregation of beta-2 microglobulin mutants: Insights from a Fourier transform infrared study in solution and in the crystalline state. Biophys. J. 2012, 102, 1676–1684. [Google Scholar] [CrossRef] [PubMed]
- Esposito, G.; Ricagno, S.; Corazza, A.; Rennella, E.; Gumral, D.; Mimmi, M.C.; Betto, E.; Pucillo, C.E.; Fogolari, F.; Viglino, P.; et al. The controlling roles of Trp60 and Trp95 in beta2-microglobulin function, folding and amyloid aggregation properties. J. Mol. Biol. 2008, 378, 887–897. [Google Scholar] [CrossRef] [PubMed]
- Camilloni, C.; Sala, B.M.; Sormanni, P.; Porcari, R.; Corazza, A.; De Rosa, M.; Zanini, S.; Barbiroli, A.; Esposito, G.; Bolognesi, M.; et al. Rational design of mutations that change the aggregation rate of a protein while maintaining its native structure and stability. Sci. Rep. 2016, 6, 25559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valleix, S.; Gillmore, J.D.; Bridoux, F.; Mangione, P.P.; Dogan, A.; Nedelec, B.; Boimard, M.; Touchard, G.; Goujon, J.M.; Lacombe, C.; et al. Hereditary systemic amyloidosis due to Asp76Asn variant beta2-microglobulin. N. Engl. J. Med. 2012, 366, 2276–2283. [Google Scholar] [CrossRef] [PubMed]
- Halabelian, L.; Ricagno, S.; Giorgetti, S.; Santambrogio, C.; Barbiroli, A.; Pellegrino, S.; Achour, A.; Grandori, R.; Marchese, L.; Raimondi, S.; et al. Class I major histocompatibility complex, the trojan horse for secretion of amyloidogenic beta2-microglobulin. J. Biol. Chem. 2014, 289, 3318–3327. [Google Scholar] [CrossRef] [PubMed]
- De Rosa, M.; Barbiroli, A.; Giorgetti, S.; Mangione, P.P.; Bolognesi, M.; Ricagno, S. Decoding the Structural Bases of D76N ss2-Microglobulin High Amyloidogenicity through Crystallography and Asn-Scan Mutagenesis. PLoS ONE 2015, 10, e0144061. [Google Scholar] [CrossRef] [PubMed]
- Myers, J.K.; Pace, C.N.; Scholtz, J.M. Denaturant m values and heat capacity changes: Relation to changes in accessible surface areas of protein unfolding. Protein Sci. 1995, 4, 2138–2148. [Google Scholar] [CrossRef]
- Le Marchand, T.; de Rosa, M.; Salvi, N.; Sala, B.M.; Andreas, L.B.; Barbet-Massin, E.; Sormanni, P.; Barbiroli, A.; Porcari, R.; Sousa Mota, C.; et al. Conformational dynamics in crystals reveal the molecular bases for D76N beta-2 microglobulin aggregation propensity. Nat. Commun. 2018, 9, 1658. [Google Scholar] [CrossRef] [Green Version]
- Mittag, T.; Forman-Kay, J.D. Atomic-level characterization of disordered protein ensembles. Curr. Opin. Struct. Biol. 2007, 17, 3–14. [Google Scholar] [CrossRef]
- McCarney, E.R.; Kohn, J.E.; Plaxco, K.W. Is there or isn’t there? The case for (and against) residual structure in chemically denatured proteins. Crit. Rev. Biochem. Mol. Biol. 2005, 40, 181–189. [Google Scholar] [CrossRef]
- Oliveberg, M.; Fersht, A.R. Thermodynamics of transient conformations in the folding pathway of barnase: Reorganization of the folding intermediate at low pH. Biochemistry 1996, 35, 2738–2749. [Google Scholar] [CrossRef] [PubMed]
- Morrone, A.; McCully, M.E.; Bryan, P.N.; Brunori, M.; Daggett, V.; Gianni, S.; Travaglini-Allocatelli, C. The denatured state dictates the topology of two proteins with almost identical sequence but different native structure and function. J. Biol. Chem. 2011, 286, 3863–3872. [Google Scholar] [CrossRef] [PubMed]
- Troilo, F.; Bonetti, D.; Toto, A.; Visconti, L.; Brunori, M.; Longhi, S.; Gianni, S. The Folding Pathway of the KIX Domain. ACS Chem Biol 2017, 12, 1683–1690. [Google Scholar] [CrossRef]
- Giri, R.; Morrone, A.; Travaglini-Allocatelli, C.; Jemth, P.; Brunori, M.; Gianni, S. Folding pathways of proteins with increasing degree of sequence identities but different structure and function. Proc. Natl. Acad. Sci. USA 2012, 109, 17772–17776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plaxco, K.W.; Gross, M. Unfolded, yes, but random? Never! Nat. Struct. Biol. 2001, 8, 659–660. [Google Scholar] [CrossRef] [PubMed]
- Religa, T.L.; Markson, J.S.; Mayor, U.; Freund, S.M.; Fersht, A.R. Solution structure of a protein denatured state and folding intermediate. Nature 2005, 437, 1053–1056. [Google Scholar] [CrossRef] [PubMed]
- Shortle, D.; Ackerman, M.S. Persistence of native-like topology in a denatured protein in 8 M urea. Science 2001, 293, 487–489. [Google Scholar] [CrossRef] [PubMed]
- Iadanza, M.G.; Silvers, R.; Boardman, J.; Smith, H.I.; Karamanos, T.K.; Debelouchina, G.T.; Su, Y.; Griffin, R.G.; Ranson, N.A.; Radford, S.E. The structure of a beta2-microglobulin fibril suggests a molecular basis for its amyloid polymorphism. Nat. Commun. 2018, 9, 4517. [Google Scholar] [CrossRef]
- Barbet-Massin, E.; Ricagno, S.; Lewandowski, J.R.; Giorgetti, S.; Bellotti, V.; Bolognesi, M.; Emsley, L.; Pintacuda, G. Fibrillar vs crystalline full-length beta-2-microglobulin studied by high-resolution solid-state NMR spectroscopy. J. Am. Chem. Soc. 2010, 132, 5556–5557. [Google Scholar] [CrossRef]
- Liberta, F.; Loerch, S.; Rennegarbe, M.; Schierhorn, A.; Westermark, P.; Westermark, G.T.; Grigorieff, N.; Fandrich, M.; Schmidt, M. Cryo-EM structure of an amyloid fibril from systemic amyloidosis. bioRxiv 2018. [Google Scholar] [CrossRef]
- Swec, P.; Lavatelli, F.; Tasaki, M.; Paissoni, C.; Rognoni, P.; Maritan, M.; Brambilla, F.; Milani, P.; Mauri, P.; Camilloni, C.; et al. Cryo-EM structure of cardiac amyloid fibrils from an immunoglobulin light chain (AL) amyloidosis patient. bioRxiv 2018. [Google Scholar] [CrossRef]




| β2m Wild Type | β2m D76N | |||
| IS1/2 (M1/2) | [GdnHCl]1/2 (M) | mvalue (kcal M−1 mol−1) | [GdnHCl]1/2 (M) | mvalue (kcal M−1 mol−1) |
| 0.16 | 2.12 ± 0.07 | 1.08 ± 0.03 | 1.82 ± 0.02 | 1.45 ± 0.04 |
| 0.42 | 2.07 ± 0.09 | 0.98 ± 0.03 | 2.13 ± 0.02 | 1.25 ± 0.03 |
| 0.57 | 2.02 ± 0.06 | 0.97 ± 0.05 | 1.95 ± 0.07 | 1.10 ± 0.03 |
| 0.79 | 2.12 ± 0.06 | 1.02 ± 0.02 | 1.79 ± 0.03 | 1.07 ± 0.03 |
| 0.91 | 2.21 ± 0.06 | 0.93 ± 0.01 | 2.03 ± 0.02 | 1.06 ± 0.04 |
| 1.01 | 1.61 ± 0.16 | 0.91 ± 0.06 | 1.42 ± 0.06 | 1.06 ± 0.06 |
| β2m Wild Type | β2m D76N | |||
| pH | [GdnHCl]1/2 (M) | mvalue (kcal M−1 mol−1) | [GdnHCl]1/2 (M) | mvalue (kcal M−1 mol−1) |
| 5.5 | <1 | 1.02 ± 0.03 | 1.22 ± 0.02 | 1.88 ± 0.04 |
| 6 | 1.40 ± 0.16 | 0.77 ± 0.04 | 1.74 ± 0.02 | 1.74 ± 0.05 |
| 7 | 2.13 ± 0.07 | 1.08 ± 0.03 | 1.81 ± 0.02 | 1.45 ± 0.04 |
| 7.5 | 2.09 ± 0.08 | 1.04 ± 0.03 | 2.36 ± 0.02 | 1.28 ± 0.03 |
| 8 | 2.04 ± 0.09 | 1.00 ± 0.03 | 1.77 ± 0.05 | 1.12 ± 0.06 |
| 8.5 | 1.95 ± 0.04 | 1.23 ± 0.01 | 1.79 ± 0.03 | 1.02 ± 0.02 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Visconti, L.; Malagrinò, F.; Broggini, L.; De Luca, C.M.G.; Moda, F.; Gianni, S.; Ricagno, S.; Toto, A. Investigating the Molecular Basis of the Aggregation Propensity of the Pathological D76N Mutant of Beta-2 Microglobulin: Role of the Denatured State. Int. J. Mol. Sci. 2019, 20, 396. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20020396
Visconti L, Malagrinò F, Broggini L, De Luca CMG, Moda F, Gianni S, Ricagno S, Toto A. Investigating the Molecular Basis of the Aggregation Propensity of the Pathological D76N Mutant of Beta-2 Microglobulin: Role of the Denatured State. International Journal of Molecular Sciences. 2019; 20(2):396. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20020396
Chicago/Turabian StyleVisconti, Lorenzo, Francesca Malagrinò, Luca Broggini, Chiara Maria Giulia De Luca, Fabio Moda, Stefano Gianni, Stefano Ricagno, and Angelo Toto. 2019. "Investigating the Molecular Basis of the Aggregation Propensity of the Pathological D76N Mutant of Beta-2 Microglobulin: Role of the Denatured State" International Journal of Molecular Sciences 20, no. 2: 396. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20020396