Effects of Iron Oxide Nanoparticles (γ-Fe2O3) on Liver, Lung and Brain Proteomes following Sub-Acute Intranasal Exposure: A New Toxicological Assessment in Rat Model Using iTRAQ-Based Quantitative Proteomics
Abstract
:1. Introduction
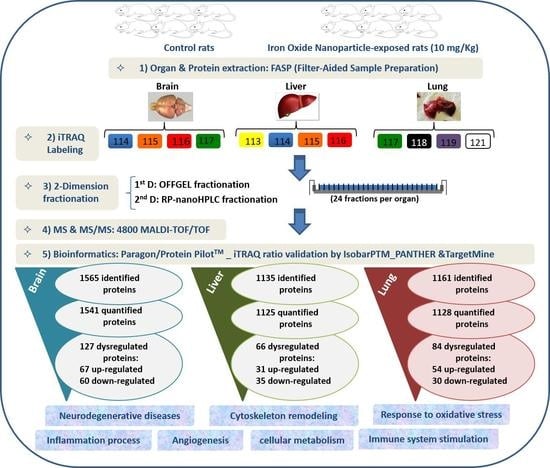
2. Results and Discussion
3. Material and Methods
3.1. Iron Oxide Nanoparticles Synthesis and Characterization
3.2. Animal Housing and Exposure Protocol
3.3. Organ/Protein Extraction and Lysis
3.4. Protein Quantification
3.5. Peptide Digestion, Desalting and iTRAQ Labelling
3.6. Peptide Separation by OFFGEL Isoelectrofocusing Followed by Reverse Phase Nano-liquid Chromatography
3.7. MALDI-TOF/TOF Analysis
3.8. Statistical Analysis of iTRAQ Data
3.9. Gene Ontology, Pathway Analysis and Protein-Protein Interaction
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Shin, S.; Song, I.; Um, S. Role of Physicochemical Properties in Nanoparticle Toxicity. Nanomaterials 2015, 5, 1351–1365. [Google Scholar] [CrossRef] [Green Version]
- Cortajarena, A.L.; Ortega, D.; Ocampo, S.M.; Gonzalez-García, A.; Couleaud, P.; Miranda, R.; Belda-Iniesta, C.; Ayuso-Sacido, A. Engineering Iron Oxide Nanoparticles for Clinical Settings. Nanobiomedicine 2014, 1, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gajanan, K.; Tijare, S.N. Applications of nanomaterials. Materials Today: Proceedings 2018, 5, 1093–1096. [Google Scholar] [CrossRef]
- Raj, S.; Sumod, U.; Jose, S.; Sabitha, M. Nanotechnology in cosmetics: Opportunities and challenges. J. Pharm. Bioallied Sci. 2012, 4, 186. [Google Scholar] [CrossRef] [PubMed]
- Stueckle, T.; Antonini, J.; Rojanasakul, Y.; Castranova, V.; Yang, Y.; Wang, L. Potential Toxicity and Underlying Mechanisms Associated with Pulmonary Exposure to Iron Oxide Nanoparticles: Conflicting Literature and Unclear Risk. Nanomaterials 2017, 7, 307. [Google Scholar] [Green Version]
- Bahadar, H.; Maqbool, F.; Niaz, K.; Abdollahi, M. Toxicity of nanoparticles and an overview of current experimental models. Iran. Biomed. J. 2016, 20, 1–11. [Google Scholar]
- Lanone, S.; Rogerieux, F.; Geys, J.; Dupont, A.; Maillot-Marechal, E.; Boczkowski, J.; Lacroix, G.; Hoet, P. Comparative toxicity of 24 manufactured nanoparticles in human alveolar epithelial and macrophage cell lines. Part. Fibre Toxicol. 2009, 6, 1–12. [Google Scholar] [CrossRef]
- Jones, C.F.; Grainger, D.W. In vitro assessments of nanomaterial toxicity. Adv. Drug Deliv. Rev. 2009, 61, 438–456. [Google Scholar] [CrossRef] [Green Version]
- Triboulet, S.; Aude-Garcia, C.; Carrière, M.; Diemer, H.; Proamer, F.; Habert, A.; Chevallet, M.; Collin-Faure, V.; Strub, J.-M.; Hanau, D.; et al. Molecular Responses of Mouse Macrophages to Copper and Copper Oxide Nanoparticles Inferred from Proteomic Analyses. Mol. Cell. Proteom. 2013, 12, 3108–3122. [Google Scholar] [CrossRef] [Green Version]
- Rahmani, S.; Mogharizadeh, L.; Attar, F.; Rezayat, S.M.; Mousavi, S.E.; Falahati, M. Probing the interaction of silver nanoparticles with tau protein and neuroblastoma cell line as nervous system models. J. Biomol. Struct. Dyn. 2018, 36, 4057–4071. [Google Scholar] [CrossRef]
- Sun, J.; Zhang, Q.; Wang, Z.; Yan, B. Effects of nanotoxicity on female reproductivity and fetal development in animal models. Int. J. Mol. Sci. 2013, 14, 9319–9337. [Google Scholar] [CrossRef] [PubMed]
- van Pomeren, M.; Peijnenburg, W.J.G.M.; Brun, N.R.; Vijver, M.G. A novel experimental and modelling strategy for nanoparticle toxicity testing enabling the use of small quantities. Int. J. Environ. Res. Public Health 2017, 14, 1348. [Google Scholar] [CrossRef] [PubMed]
- Villacis, R.A.R.; Filho, J.S.; Piña, B.; Azevedo, R.B.; Pic-Taylor, A.; Mazzeu, J.F.; Grisolia, C.K. Integrated assessment of toxic effects of maghemite (γ-Fe 2 O 3 ) nanoparticles in zebrafish. Aquat. Toxicol. 2017, 191, 219–225. [Google Scholar] [CrossRef] [PubMed]
- Brohi, R.D.; Wang, L.; Talpur, H.S.; Wu, D.; Khan, F.A.; Bhattarai, D.; Rehman, Z.U.; Farmanullah, F.; Huo, L.J. Toxicity of nanoparticles on the reproductive system in animal models: A review. Front. Pharmacol. 2017, 8, 1–22. [Google Scholar] [CrossRef]
- Chakraborty, C.; Sharma, A.R.; Sharma, G.; Lee, S.S. Zebrafish: A complete animal model to enumerate the nanoparticle toxicity. J. Nanobiotechnol. 2016, 14, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Qin, Z.; Zeng, W.; Yang, T.; Cao, Y.; Mei, C.; Kuang, Y. Toxicity assessment of nanoparticles in various systems and organs. Nanotechnol. Rev. 2017, 6, 279–289. [Google Scholar] [CrossRef]
- González-Flecha, B. Oxidant mechanisms in response to ambient air particles. Mol. Aspects Med. 2004, 25, 169–182. [Google Scholar] [CrossRef]
- Wolf-Grosse, S.; Rokstad, A.M.; Ali, S.; Lambris, J.D.; Mollnes, T.E.; Nilsen, A.M.; Stenvik, J. Iron oxide nanoparticles induce cytokine secretion in a complement-dependent manner in a human whole blood model. Int. J. Nanomedicine 2017, 12, 3927–3940. [Google Scholar] [CrossRef]
- Khan, M.I.; Mohammad, A.; Patil, G.; Naqvi, S.A.H.; Chauhan, L.K.S.; Ahmad, I. Induction of ROS, mitochondrial damage and autophagy in lung epithelial cancer cells by iron oxide nanoparticles. Biomaterials 2012, 33, 1477–1488. [Google Scholar] [CrossRef]
- Alarifi, S.; Ali, D.; Alkahtani, S.; Alhader, M.S. Iron oxide nanoparticles induce oxidative stress, DNA damage, and caspase activation in the human breast cancer cell line. Biol. Trace Elem. Res. 2014, 159, 416–424. [Google Scholar] [CrossRef]
- Wang, L.; Wang, L.; Ding, W.; Zhang, F. Acute Toxicity of Ferric Oxide and Zinc Oxide Nanoparticles in Rats. J. Nanosci. Nanotechnol. 2010, 10, 8617–8624. [Google Scholar] [CrossRef] [PubMed]
- Sadeghi, L.; Yousefi Babadi, V.; Espanani, H.R. Toxic effects of the Fe2O3 nanoparticles on the liver and lung tissue. Bratisl. Lek. Listy 2015, 116, 373–378. [Google Scholar] [CrossRef] [PubMed]
- Van Rooy, I.; Cakir-Tascioglu, S.; Hennink, W.E.; Storm, G.; Schiffelers, R.M.; Mastrobattista, E. In vivo methods to study uptake of nanoparticles into the brain. Pharm. Res. 2011, 28, 456–471. [Google Scholar] [CrossRef] [PubMed]
- Rabilloud, T.; Lescuyer, P. Proteomics in mechanistic toxicology: History, concepts, achievements, caveats, and potential. Proteomics 2015, 15, 1051–1074. [Google Scholar] [CrossRef] [PubMed]
- Matysiak, M.; Kapka-Skrzypczak, L.; Brzóska, K.; Gutleb, A.C.; Kruszewski, M. Proteomic approach to nanotoxicity. J. Proteomics 2016, 137, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Fröhlich, E. Role of omics techniques in the toxicity testing of nanoparticles. J. Nanobiotechnol. 2017, 15, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Nath Roy, D.; Goswami, R.; Pal, A. Nanomaterial and toxicity: What can proteomics tell us about the nanotoxicology? Xenobiotica 2017, 47, 632–643. [Google Scholar] [CrossRef]
- Lehmann, S.G.; Bourgoin-Voillard, S.; Seve, M.; Rachidi, W. Tubulin Beta-3 Chain as a New Candidate Protein Biomarker of Human Skin Aging: A Preliminary Study. Oxid. Med. Cell. Longev. 2017, 2017, 5140360. [Google Scholar] [CrossRef]
- Lehmann, S.G.; Seve, M.; Vanwonterghem, L.; Michelland, S.; Cunin, V.; Coll, J.L.; Hurbin, A.; Bourgoin-Voillard, S. A large scale proteome analysis of the gefitinib primary resistance overcome by KDAC inhibition in KRAS mutated adenocarcinoma cells overexpressing amphiregulin. J. Proteomics 2019, 195, 114–124. [Google Scholar] [CrossRef]
- Askri, D.; Cunin, V.; Béal, D.; Berthier, S.; Chovelon, B.; Arnaud, J.; Rachidi, W.; Sakly, M.; Amara, S.; Sève, M.; et al. Investigating the toxic effects induced by iron oxide nanoparticles on neuroblastoma cell line: An integrative study combining cytotoxic, genotoxic and proteomic tools. Nanotoxicology 2019, 1–20. [Google Scholar] [CrossRef]
- Askri, D.; Ouni, S.; Galai, S.; Arnaud, J.; Chovelon, B.; Lehmann, S.G.; Sturm, N.; Sakly, M.; Sève, M.; Amara, S. Intranasal instillation of iron oxide nanoparticles induces inflammation and perturbation of trace elements and neurotransmitters, but not behavioral impairment in rats. Environ. Sci. Pollut. Res. 2018, 25, 16922–16932. [Google Scholar] [CrossRef] [PubMed]
- Oberdörster, G.; Sharp, Z.; Atudorei, V.; Elder, A.; Gelein, R.; Kreyling, W.; Cox, C. Translocation of inhaled ultrafine particles to the brain. Inhal. Toxicol. 2004, 16, 437–445. [Google Scholar]
- Mistry, A.; Stolnik, S.; Illum, L. Nose-to-Brain Delivery: Investigation of the Transport of Nanoparticles with Different Surface Characteristics and Sizes in Excised Porcine Olfactory Epithelium. Mol. Pharm. 2015, 12, 2755–2766. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.A.; Tripathi, L.P.; Mizuguchi, K. TargetMine, an integrated data warehouse for candidate gene prioritisation and target discovery. PLoS ONE 2011, 6, e17844. [Google Scholar] [CrossRef]
- Mi, H.; Muruganujan, A.; Thomas, P.D. PANTHER in 2013: Modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2013, 41, 377–386. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Fahmy, B.; Cormier, S. Copper oxide nanoparticles induce oxidative stress and cytotoxicity in airway epithelial cells. Toxicol. Vitr. 2009, 23, 1365–1371. [Google Scholar] [CrossRef] [Green Version]
- Khalaf, A.A.; Zaki, A.R.; Galal, M.K.; Ogaly, H.A.; Ibrahim, M.A.; Hassan, A. The potential protective effect of α-lipoic acid against nanocopper particle-induced hepatotoxicity in male rats. Hum. Exp. Toxicol. 2017, 36, 881–891. [Google Scholar] [CrossRef]
- Ribeiro, M.J.; Maria, V.L.; Scott-Fordsmand, J.J.; Amorim, M.J.B. Oxidative stress mechanisms caused by Ag nanoparticles (NM300K) are different from those of AgNO3: Effects in the soil invertebrate Enchytraeus Crypticus. Int. J. Environ. Res. Public Health 2015, 12, 9589–9602. [Google Scholar] [CrossRef]
- Paszek, E.; Czyz, J.; Woźnicka, O.; Jakubiak, D.; Wojnarowicz, J.; Łojkowski, W.; Stȩpień, E. Zinc oxide nanoparticles impair the integrity of human umbilical vein endothelial cell monolayer in vitro. J. Biomed. Nanotechnol. 2012, 8, 957–967. [Google Scholar] [CrossRef]
- Gaiser, B.K.; Hirn, S.; Kermanizadeh, A.; Kanase, N.; Fytianos, K.; Wenk, A.; Haberl, N.; Brunelli, A.; Kreyling, W.G.; Stone, V. Effects of silver nanoparticles on the liver and hepatocytes in vitro. Toxicol. Sci. 2013, 131, 537–547. [Google Scholar] [CrossRef] [PubMed]
- Soenen, S.J.H.; Nuytten, N.; De Meyer, S.F.; De Smedt, S.C.; De Cuyper, M. High intracellular iron oxide nanoparticle concentrations affect cellular cytoskeleton and focal adhesion kinase-mediated signaling. Small 2010, 6, 832–842. [Google Scholar] [CrossRef] [PubMed]
- Hoskins, C.; Cuschieri, A.; Wang, L. The cytotoxicity of polycationic iron oxide nanoparticles: Common endpoint assays and alternative approaches for improved understanding of cellular response mechanism. J. Nanobiotechnol. 2012, 10, 15. [Google Scholar] [CrossRef] [PubMed]
- Triboulet, S.; Aude-Garcia, C.; Armand, L.; Collin-Faure, V.; Chevallet, M.; Diemer, H.; Gerdil, A.; Proamer, F.; Strub, J.M.; Habert, A.; et al. Comparative proteomic analysis of the molecular responses of mouse macrophages to titanium dioxide and copper oxide nanoparticles unravels some toxic mechanisms for copper oxide nanoparticles in macrophages. PLoS ONE 2015, 10, e0124496. [Google Scholar] [CrossRef] [PubMed]
- Hachani, R.; Birchall, M.A.; Lowdell, M.W.; Kasparis, G.; Tung, L.D.; Manshian, B.B.; Soenen, S.J.; Gsell, W.; Himmelreich, U.; Gharagouzloo, C.A.; et al. Assessing cell-nanoparticle interactions by high content imaging of biocompatible iron oxide nanoparticles as potential contrast agents for magnetic resonance imaging. Sci. Rep. 2017, 7, 7850. [Google Scholar] [CrossRef]
- Panariti, A.; Miserocchi, G.; Rivolta, I. The effect of nanoparticle uptake on cellular behavior: Disrupting or enabling functions? Nanotechnol. Sci. Appl. 2012, 5, 87–100. [Google Scholar]
- Buyukhatipoglu, K.; Clyne, A.M. Superparamagnetic iron oxide nanoparticles change endothelial cell morphology and mechanics via reactive oxygen species formation. J. Biomed. Mater. Res. Part A 2011, 96, 186–195. [Google Scholar] [CrossRef]
- Pollak, D.; Cairns, N.; Lubec, G. Cytoskeleton derangement in brain of patients with Down Syndrome, Alzheimer’s disease and Pick’s disease. In Advances in Down Syndrome Research; Springer: Vienna, Austria, 2003. [Google Scholar]
- Räisänen, S.R.; Lehenkari, P.; Tasanen, M.; Rahkila, P.; Härkönen, P.L.; Väänänen, H.K. Carbonic anhydrase III protects cells from hydrogen peroxide-induced apoptosis. FASEB J. 1999, 13, 513–522. [Google Scholar] [CrossRef]
- Shi, C.; Uda, Y.; Dedic, C.; Azab, E.; Sun, N.; Hussein, A.I.; Petty, C.A.; Fulzele, K.; Mitterberger-Vogt, M.C.; Zwerschke, W.; et al. Carbonic anhydrase III protects osteocytes from oxidative stress. FASEB J. 2018, 32, 440–452. [Google Scholar] [CrossRef]
- Da Fonseca, R.R.; Johnson, W.E.; O’Brien, S.J.; Vasconcelos, V.; Antunes, A. Molecular evolution and the role of oxidative stress in the expansion and functional diversification of cytosolic glutathione transferases. BMC Evol. Biol. 2010, 10, 281. [Google Scholar] [CrossRef]
- Klaus, A.; Zorman, S.; Berthier, A.; Polge, C.; Ramirez, S.; Michelland, S.; Sève, M.; Vertommen, D.; Rider, M.; Lentze, N.; et al. Glutathione S-Transferases Interact with AMP-Activated Protein Kinase: Evidence for S-Glutathionylation and Activation In Vitro. PLoS ONE 2013, 8, e62497. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-R.; Kuo, C.-J.; Lin, H.Y.-H.; Wu, C.-J.; Liang, S.-S. A Proteomics Analysis to Evaluate Cytotoxicity in NRK-52E Cells Caused by Unmodified Nano-Fe3O4. Sci. World J. 2014, 2014, 1–9. [Google Scholar] [CrossRef]
- Singhal, A.; Morris, V.B.; Labhasetwar, V.; Ghorpade, A. Nanoparticle-mediated catalase delivery protects human neurons from oxidative stress. Cell Death Dis. 2013, 4, e903–e909. [Google Scholar] [CrossRef] [PubMed]
- Gutch, M.; Consil, S.; Kumar, S.; Agarwal, A.; Robo, R.; Jain, N. Oxidative stress determined through the levels of antioxidant enzymes and the effect of N-acetylcysteine in aluminum phosphide poisoning. Indian J. Crit. Care Med. 2014, 18, 666. [Google Scholar]
- Gautam, A.; Ray, A.; Mukherjee, S.; Das, S.; Pal, K.; Das, S.; Karmakar, P.; Ray, M.; Ray, S. Immunotoxicity of copper nanoparticle and copper sulfate in a common Indian earthworm. Ecotoxicol. Environ. Saf. 2018, 148, 620–631. [Google Scholar] [CrossRef]
- Kim, S.J.; Yune, T.Y.; Han, C.T.; Kim, Y.C.; Oh, Y.J.; Markelonis, G.J.; Oh, T.H. Mitochondrial isocitrate dehydrogenase protects human neuroblastoma SH-SY5Y cells against oxidative stress. J. Neurosci. Res. 2007, 85, 139–152. [Google Scholar] [CrossRef]
- Lee, S.H.; Ha, S.O.; Koh, H.J.; Kim, K.; Jeon, S.M.; Choi, M.S.; Kwon, O.S.; Huh, T.L. Upregulation of cytosolic NADP+-dependent isocitrate dehydrogenase by hyperglycemia protects renal cells against oxidative stress. Mol. Cells 2010, 29, 203–208. [Google Scholar] [CrossRef]
- Wang, P.-F.; Song, H.-W.; Cai, H.-Q.; Kong, L.-W.; Yao, K.; Jiang, T.; Li, S.-W.; Yan, C.-X. Preoperative inflammation markers and IDH mutation status predict glioblastoma patient survival. Oncotarget 2017, 8, 50117–50123. [Google Scholar] [CrossRef] [Green Version]
- Dalli, J.; Norling, L.V.; Renshaw, D.; Cooper, D.; Leung, K.; Perretti, M. Annexin 1 mediates the rapid anti-inflammatory effects of neutrophil-derived microparticles. Blood 2018, 112, 2512–2520. [Google Scholar] [CrossRef]
- Mirsaeidi, M.; Gidfar, S.; Vu, A.; Schraufnagel, D. Annexins family: Insights into their functions and potential role in pathogenesis of sarcoidosis. J. Transl. Med. 2016, 14, 1–9. [Google Scholar] [CrossRef]
- Zhao, B.; Wang, J.; Liu, L.; Li, X.; Liu, S.; Xia, Q.; Shi, J. Annexin A1 translocates to nucleus and promotes the expression of pro-inflammatory cytokines in a PKC-dependent manner after OGD/R. Sci. Rep. 2016, 6, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Gilthorpe, J.D.; Oozeer, F.; Nash, J.; Calvo, M.; Bennett, D.L.; Lumsden, A.; Pini, A. Extracellular histone H1 is neurotoxic and drives a pro-inflammatory response in microglia. F1000Research 2013, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Kang, R.; Fan, X.-G.; Tang, D. Release and activity of histone in diseases. Cell Death Dis. 2014, 5, e1370. [Google Scholar] [CrossRef] [PubMed]
- Vogl, T.; Gharibyan, A.L.; Morozova-Roche, L.A. Pro-inflammatory S100A8 and S100A9 proteins: Self-assembly into multifunctional native and amyloid complexes. Int. J. Mol. Sci. 2012, 13, 2893–2917. [Google Scholar] [CrossRef] [PubMed]
- Lemine, O.M.; Omri, K.; Iglesias, M.; Velasco, V.; Crespo, P.; De La Presa, P.; El Mir, L.; Bouzid, H.; Yousif, A.; Al-Hajry, A. ??-Fe2O3by sol-gel with large nanoparticles size for magnetic hyperthermia application. J. Alloys Compd. 2014, 607, 125–131. [Google Scholar] [CrossRef]
- Wisniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M.; Wi, J.R. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Campone, M.; Valo, I.; Jézéquel, P.; Moreau, M.; Boissard, A.; Campion, L.; Loussouarn, D.; Verriele, V.; Coqueret, O.; Guette, C. Prediction of Recurrence and Survival for Triple-Negative Breast Cancer (TNBC) by a Protein Signature in Tissue Samples. Mol. Cell. Proteomics 2015, 14, 2936–2946. [Google Scholar] [CrossRef] [Green Version]
- Mi, H.; Muruganujan, A.; Ebert, D.; Huang, X.; Thomas, P.D. PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019, 47, D419–D426. [Google Scholar] [CrossRef]






| Pathway (Pathway Accession) | Number of Proteins | Percentage % (from 66 Proteins) | Protein Accession Number | Protein Name |
|---|---|---|---|---|
| Pyruvate metabolism (P02772) | 2 | 3.1% | P12928 | Pyruvate kinase PKLR |
| P16638 | ATP-citrate synthase | |||
| Blood coagulation (P00011) | 1 | 1.5% | P02680 | Fibrinogen gamma chain |
| Pentose phosphate pathway (P02762) | 1 | 1.5% | P50137 | Transketolase |
| Huntington disease (P00029) | 3 | 4.6% | P04797 | Glyceraldehyde-3-phosphate dehydrogenase |
| P61206 | ADP-ribosylation factor 3 | |||
| D3ZFQ8 | Cytochrome c-1 | |||
| Arginine biosynthesis (P02728) | 1 | 1.5% | P09034 | Argininosuccinate synthase |
| Glycolysis (P00024) | 2 | 3.1% | P04797 | Glyceraldehyde-3-phosphate dehydrogenase |
| P12928 | Pyruvate kinase | |||
| Parkinson disease (P00049) | 1 | 1.5% | P63102 | 14-3-3 protein zeta/delta |
| Plasminogen activating cascade (P00050) | 1 | 1.5% | P02680 | Fibrinogen gamma chain |
| PI3 kinase pathway (P00048) | 1 | 1.5% | P63102 | 14-3-3 protein zeta/delta |
| FGF signaling pathway (P00021) | 1 | 1.5% | P63102 | 14-3-3 protein zeta/delta |
| ATP synthesis (P02721) | 1 | 1.5% | D3ZFQ8 | Cytochrome c-1 |
| Serine glycine biosynthesis (P02776) | 1 | 1.5% | Q5U3Z7 | Serine hydroxymethyltransferase |
| EGF receptor signaling pathway (P00018) | 1 | 1.5% | P63102 | 14-3-3 protein zeta/delta |
| FAS signaling pathway (P00020) | 1 | 1.5% | D3ZFQ8 | Cytochrome c-1 |
| Pathway (Pathway Accession) | Number of Proteins | Percentage % (from 127 Proteins) | Protein Accession Number | Protein Name |
|---|---|---|---|---|
| 5HT1 type receptor mediated signaling pathway (P04373) | 1 | 0.8 | P27791 | cAMP-dependent protein kinase catalytic subunit alpha |
| Alzheimer disease-amyloid secretase pathway (P00003) | 2 | 1.6 | P08592 | Amyloid-beta A4 protein |
| P49186 | Mitogen-activated protein kinase 9 | |||
| Beta1 adrenergic receptor signaling pathway (P04377) | 1 | 0.8 | P27791 | cAMP-dependent protein kinase catalytic subunit alpha |
| Beta2 adrenergic receptor signaling pathway (P04378) | 1 | 0.8 | P27791 | cAMP-dependent protein kinase catalytic subunit alpha |
| Dopamine receptor mediated signaling pathway (P05912) | 3 | 2.4 | Q6J4I0 | Protein phosphatase 1 regulatory subunit 1B |
| P27791 | cAMP-dependent protein kinase catalytic subunit alpha | |||
| P19627 | Guanine nucleotide-binding protein G(z) subunit alpha | |||
| Muscarinic acetylcholine receptor 2 and 4 signaling pathway | 1 | 0.8 | P27791 | cAMP-dependent protein kinase catalytic subunit alpha |
| Enkephalin release (P05913) | 1 | 0.8 | P27791 | cAMP-dependent protein kinase catalytic subunit alpha |
| GABA-B receptor II signaling (P05731) | 1 | 0.8 | P27791 | cAMP-dependent protein kinase catalytic subunit alpha |
| Gamma-aminobutyric acid synthesis (P04384) | 1 | 0.8 | P51650 | Succinate-semialdehyde dehydrogenase, mitochondrial |
| Gonadotropin-releasing hormone receptor pathway (P6664) | 3 | 2.4 | P49186 | Mitogen-activated protein kinase 9 |
| P14668 | Annexin A5 | |||
| P63055 | Calmodulin regulator protein PCP4 | |||
| Heterotrimeric G-protein signaling pathway-Gi alpha and Gs alpha mediated pathway (P00026) | 1 | 0.8 | P27791 | cAMP-dependent protein kinase catalytic subunit alpha |
| Heterotrimeric G-protein signaling pathway-rod outer segment phototransduction (P00028) | 1 | 0.8 | P27791 | cAMP-dependent protein kinase catalytic subunit alpha |
| Metabotropic glutamate receptor group I pathway (P00041) | 1 | 0.8 | P27791 | cAMP-dependent protein kinase catalytic subunit alpha |
| Metabotropic glutamate receptor group II pathway (P00040) | 1 | 0.8 | P27791 | cAMP-dependent protein kinase catalytic subunit alpha |
| Metabotropic glutamate receptor group III pathway (P00039) | 1 | 0.8 | P27791 | cAMP-dependent protein kinase catalytic subunit alpha |
| Huntington disease (P00029) | 4 | 3.2 | P49186 | Mitogen-activated protein kinase 9 |
| Q3KRE8 | Tubulin beta-2B chain | |||
| P85108 | Tubulin beta-2A chain | |||
| Q07009 | Calpain-2 catalytic subunit | |||
| Nicotine pharmacodynamics pathway (P06587) | 2 | 1.6 | Q6J4I0 | Protein phosphatase 1 regulatory subunit 1B |
| P27791 | cAMP-dependent protein kinase catalytic subunit alpha | |||
| Nicotinic acetylcholine receptor signaling pathway (P00044) | 1 | 0.8 | Q62812 | Myosin-9 |
| Parkinson disease (P00049) | 2 | 1.6 | P49186 | Mitogen-activated protein kinase 9 |
| P37377 | Alpha-synuclein |
| Pathway (Pathway Accession) | Number of Proteins | Percentage % (from 127 Proteins) | Protein Accession Number | Protein Name |
|---|---|---|---|---|
| Apoptosis signaling pathway (P00006) | 1 | 0.8 | P49186 | Mitogen-activated protein kinase 9 |
| Toll receptor signaling pathway (P00054) | 1 | 0.8 | P49186 | Mitogen-activated protein kinase 9 |
| B cell activation (P00010) | 1 | 0.8 | P49186 | Mitogen-activated protein kinase 9 |
| T cell activation (P00053) | 1 | 0.8 | P49186 | Mitogen-activated protein kinase 9 |
| Oxidative stress response (P00046) | 1 | 0.8 | P49186 | Mitogen-activated protein kinase 9 |
| Inflammation mediated by chemokine and cytokine signaling pathway (P00031) | 2 | 1.6 | P49186 | Mitogen-activated protein kinase 9 |
| P27791 | cAMP-dependent protein kinase catalytic subunit alpha | |||
| Integrin signaling pathway (P00034) | 1 | 0.8 | P49186 | Mitogen-activated protein kinase 9 |
| Interferon-gamma signaling pathway (P00035) | 1 | 0.8 | P49186 | Mitogen-activated protein kinase 9 |
| Histamine H2 receptor mediated signaling pathway (P04386) | 1 | 0.8 | P27791 | cAMP-dependent protein kinase catalytic subunit alpha |
| Ras Pathway (P04393) | 1 | 0.8 | P49186 | Mitogen-activated protein kinase 9 |
| CCKR signaling map (P06959) | 3 | 2.4 | P49186 | Mitogen-activated protein kinase 9 |
| P13234 | Calcium/calmodulin-dependent protein kinase type IV | |||
| P27791 | cAMP-dependent protein kinase catalytic subunit alpha | |||
| Cytoskeletal regulation by Rho GTPase (P00016) | 3 | 2.4 | Q3KRE8 | Tubulin beta-2B chain |
| Q62812 | Myosin-9 | |||
| P85108 | Tubulin beta-2A chain | |||
| De novo purine biosynthesis (P02738) | 2 | 1.6 | O35567 | Bifunctional purine biosynthesis protein PURH |
| P19804 | Nucleoside diphosphate kinase B | |||
| De novo pyrimidine deoxyribonucleotide biosynthesis (P02739) | 1 | 0.8 | P19804 | Nucleoside diphosphate kinase B |
| De novo pyrimidine ribonucleotides biosythesis (P02740) | 1 | 0.8 | P19804 | Nucleoside diphosphate kinase B |
| Biological Process | Accession Number | Ratio Exp/Ctr | p-Value |
|---|---|---|---|
| Response to oxidative stress | |||
| Glutathione S-transferase Yb-3 | P08009 | 1.62 | <0.001 |
| NADH dehydrogenase | B0BNE6 | 2.30 | 0.027 |
| Inflammation process | |||
| Annexin A5 | P14668 | 1.37 | 0.02 |
| Cancer: angio/lymphangiogenesis | |||
| Ezrin | P31977 | 1.31 | 0.0038 |
| Malignant T-cell-amplified sequence 1 | Q4G009 | 1.30 | 0.012 |
| Neuronal damage related to Alzheimer Disease | |||
| Neuronal pentraxin-1 | P47971 | 1.36 | 0.009 |
| Adherence, neuronal differentiation and signal transduction | |||
| Disks large homolog 1 | Q62696 | 0.56 | 0.025 |
| Cell cycle exit and neuronal differentiation protein1 | Q5FVI4 | 0.73 | 0.015 |
| Biological Process | Accession Number | Ratio Exp/Ctr | p-Value |
|---|---|---|---|
| Response to oxidative stress | |||
| Carbonic anhydrase 3 | P14141 | 2.07 | <0.001 |
| Glutathione S-transferase alpha-1 | P00502 | 1.30 | <0.001 |
| Glutathione S-transferase Mu 2 | P08010 | 1.30 | 0.005 |
| Catalase | P04762 | 1.26 | 0.015 |
| Isocitrate dehydrogenase | P41562 | 1.20 | 0.012 |
| Immune system | |||
| Ig gamma-1 chain C region | P20759 | 1.56 | 0.029 |
| Cancer metabolism | |||
| Sulfotransferase 1C1 | P50237 | 1.66 | <0.001 |
| Hepatic lipid metabolism | |||
| Vigilin | Q9Z1A6 | 1.47 | 0.012 |
| Fatty Acide Metabolism | |||
| Long-chain-fatty-acid--CoA ligase 5 | O88813 | 0.73 | 0.022 |
| Fatty acid synthase | P12785 | 0.63 | <0.001 |
| Glycolysis/Gluconeogenesis | |||
| ATP-citrate synthase | P16638 | 0.59 | <0.001 |
| Glyceraldehyde-3-phosphate dehydrogenase | P04797 | 0.79 | 0.025 |
| Glucose-6-phosphate 1-dehydrogenase | P05370 | 0.60 | 0.008 |
| Pyruvate kinase | P12928 | 0.66 | <0.001 |
| Cytochrome C1 | D3ZFQ8 | 0.82 | 0.043 |
| Oxidative Phosphorylation | |||
| Cytochrome c oxidase COX 7B | P80431 | 0.65 | 0.046 |
| Biological Process | Accession Number | Ratio Exp/ctr | p-Value |
|---|---|---|---|
| Response to oxidative stress | |||
| Superoxide dismutase | P07895 | 1.48 | 0.005 |
| Peroxiredoxin-6 | O35244 | 1.26 | 0.049 |
| Redox-regulatory protein FAM213A | Q6AXX6 | 1.22 | 0.013 |
| Immune system stimulation and response to inflammation | |||
| Annexin | Q5XI77 | 1.54 | 0.021 |
| Histone H1.5 | D3ZBN0 | 1.35 | 0.025 |
| Histone H2A | Q6I8Q6 | 1.75 | 0.021 |
| Protein S100-A8 | P50115 | 1.30 | 0.009 |
| Protein S100-A9 | P50116 | 1.65 | <0.001 |
| Receptor-type tyrosine-protein phosphatase C | P04157 | 1.71 | 0.004 |
| Redox-regulatory protein FAM213A | Q6AXX6 | 1.22 | 0.013 |
| Cell death | |||
| Programmed cell death 6 | G3V7W1 | 1.23 | 0.045 |
| Iron metabolism | |||
| Ferritin heavy chain | P19132 | 1.36 | <0.001 |
| Cell adhesion and cancer progress | |||
| Podocalyxin | Q9WTQ2 | 1.96 | 0.002 |
| Metabolism | |||
| Succinate dehydrogenase | P21913 | 0.71 | 0.005 |
| Phosphoglycerate kinase 1 | P16617 | 0.79 | 0.044 |
| Oxygen transport (lung to tissues) | |||
| Hemoglobin subunit alpha-1/2 | P01946 | 0.37 | <0.001 |
| Hemoglobin subunit beta-1 and beta-2 | P02091 | 0.77 | <0.001 |
| Hemostasis | |||
| Fibrinogen gamma chain | P02680 | 0.79 | 0.008 |
| Fibrinogen beta chain | P14480 | 0.81 | 0.017 |
| Cell proliferation and differentiation | |||
| Thymosin beta-4 | P62329 | 0.61 | <0.001 |
| Organ | Accession Number | Protein Name | Panther Protein Class |
|---|---|---|---|
| Brain | Q3KRE8 | Tubulin beta-2B chain | Tubulin |
| P31977 | Ezrin | Actin family cytoskeletal protein | |
| Q63610 | Tropomyosin alpha-3 chain | Actin binding motor protein | |
| Q6AY56 | Tubulin alpha-8 chain | Tubulin | |
| P13234 | Calcium/calmodulin-dependent protein kinase type IV | Non-motor microtubule binding protein Non-receptor serine/threonine protein kinase | |
| P85108 | Tubulin beta-2A chain | Tubulin | |
| Liver | Q7M0E3 | Destrin | Non-motor actin binding protein |
| P63029 | Translationally-controlled tumor protein | Ton-motor actin binding protein | |
| P09495 | Tropomyosin alpha-4 chain | Actin binding motor protein | |
| Lung | P47875 | Cysteine and glycine-rich protein 1 | Actin family cytoskeletal protein |
| Q63598 | Plastin-3 | Non-motor actin binding protein | |
| Q63355 | Unconventional myosin-Ic | G-protein modulatoractin binding motor proteincell junction protein |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Askri, D.; Cunin, V.; Ouni, S.; Béal, D.; Rachidi, W.; Sakly, M.; Amara, S.; Lehmann, S.G.; Sève, M. Effects of Iron Oxide Nanoparticles (γ-Fe2O3) on Liver, Lung and Brain Proteomes following Sub-Acute Intranasal Exposure: A New Toxicological Assessment in Rat Model Using iTRAQ-Based Quantitative Proteomics. Int. J. Mol. Sci. 2019, 20, 5186. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20205186
Askri D, Cunin V, Ouni S, Béal D, Rachidi W, Sakly M, Amara S, Lehmann SG, Sève M. Effects of Iron Oxide Nanoparticles (γ-Fe2O3) on Liver, Lung and Brain Proteomes following Sub-Acute Intranasal Exposure: A New Toxicological Assessment in Rat Model Using iTRAQ-Based Quantitative Proteomics. International Journal of Molecular Sciences. 2019; 20(20):5186. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20205186
Chicago/Turabian StyleAskri, Dalel, Valérie Cunin, Souhir Ouni, David Béal, Walid Rachidi, Mohsen Sakly, Salem Amara, Sylvia G. Lehmann, and Michel Sève. 2019. "Effects of Iron Oxide Nanoparticles (γ-Fe2O3) on Liver, Lung and Brain Proteomes following Sub-Acute Intranasal Exposure: A New Toxicological Assessment in Rat Model Using iTRAQ-Based Quantitative Proteomics" International Journal of Molecular Sciences 20, no. 20: 5186. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20205186