Expression, Subcellular Localization, and Interactions of CPK Family Genes in Maize
Abstract
:1. Introduction
2. Results
2.1. Expression Analysis of ZmCPK Family Members
2.2. Subcellular Localization of Selected ZmCPK Proteins
2.3. Interaction Between ZmCPK Family and A Subclass ZmPP2C Family
2.4. Interaction of the ZmCPK Family and the ZmSnRK2 Family
2.5. Physical Interaction Between ZmCPK and ZmMPK Gene Families
3. Discussion
3.1. Expression Profiles of ZmCPKs
3.2. Sub-Cellular Localization of CPK Genes in Maize
3.3. Interaction Analysis of ZmCPKs, ZmPP2Cs, and ZmSnRKs
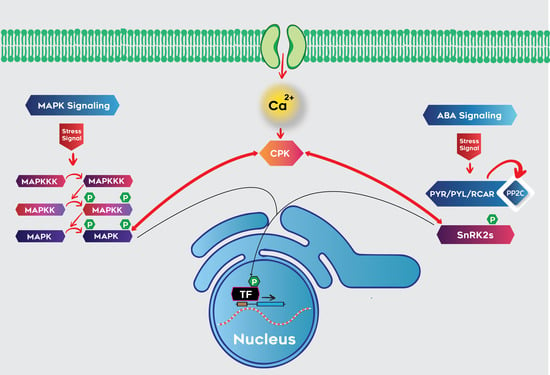
3.4. Interaction of Calcium and MAPK Signaling Pathways
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. ABA/CaCl2 Treatment and RNA Extraction
4.3. Quantitative Real-Time PCR
4.4. Subcellular Localization
4.5. Yeast Two‑Hybrid Assay
4.6. Bimolecular Fluorescence Complementation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zhu, J.K. Plant salt tolerance. Trends Plant Sci. 2001, 6, 66–71. [Google Scholar] [CrossRef]
- Xiong, L.; Schumaker, K.S.; Zhu, J.K. Cell signaling during cold, drought, and salt stress. Plant Cell 2002, 14, S165–S183. [Google Scholar] [CrossRef] [Green Version]
- Zhu, J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evans, N.H.; McAinsh, M.R.; Hetherington, A.M. Calcium oscillations in higher plants. Curr. Opin. Plant Biol. 2001, 4, 415–420. [Google Scholar] [CrossRef]
- Tuteja, N.; Mahajan, S. Calcium signaling network in plants: An overview. Plant Signal. Behav. 2007, 2, 79–85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kudla, J.; Batistič, O.; Hashimoto, K. Calcium signals: The lead currency of plant information processing. Plant Cell 2010, 22, 541–563. [Google Scholar] [CrossRef] [PubMed]
- Saijo, Y.; Hata, S.; Kyozuka, J.; Shimamoto, K.; Izui, K. Over-expression of a single Ca2+-dependent protein kinase confers both cold and salt/drought tolerance on rice plants. Plant J. 2000, 23, 319–327. [Google Scholar] [CrossRef]
- Cheng, S.H.; Willmann, M.R.; Chen, H.C.; Sheen, J. Calcium Signaling through Protein Kinases. The Arabidopsis Calcium-Dependent Protein Kinase Gene Family. Plant Physiol. 2002, 129, 469–485. [Google Scholar] [CrossRef] [Green Version]
- Harper, J.F.; Breton, G.; Harmon, A. Decoding Ca2+ signals through plant protein kinases. Annu. Rev. Plant Biol. 2004, 55, 263–288. [Google Scholar] [CrossRef]
- Xiong, T.C.; Bourque, S.; Lecourieux, D.; Amelot, N.; Grat, S.; Brière, C.; Mazars, C.; Pugin, A.; Ranjeva, R. Calcium signaling in plant cell organelles delimited by a double membrane. Biochim. Biophys. Acta Mol. Cell Res. 2006, 1763, 1209–1215. [Google Scholar] [CrossRef] [Green Version]
- Zhu, S.Y.; Yu, X.C.; Wang, X.J.; Rui, Z.; Yan, L.; Fan, R.C.; Yi, S.; Du, S.Y.; Wang, X.F.; Wu, F.Q.; et al. Two Calcium-Dependent Protein Kinases, CPK4 and CPK11, Regulate Abscisic Acid Signal Transduction in Arabidopsis. Plant Cell 2007, 19, 3019–3036. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehlmer, N.; Wurzinger, B.; Stael, S.; Hofmannrodrigues, D.; Csaszar, E.; Pfister, B.; Bayer, R.; Teige, M. The Ca2+-dependent protein kinase CPK3 is required for MAPK-independent salt-stress acclimation in Arabidopsis. Plant J. 2010, 63, 484–498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asano, T.; Hakata, M.; Nakamura, H.; Aoki, N.; Komatsu, S.; Ichikawa, H.; Hirochika, H.; Ohsugi, R. Functional characterisation of OsCPK21, a calcium-dependent protein kinase that confers salt tolerance in rice. Plant Mol. Biol. 2011, 75, 179–191. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, M.; Yoshioka, M.; Asai, S.; Nomura, H.; Kuchimura, K.; Mori, H.; Doke, N.; Yoshioka, H. StCDPK5 confers resistance to late blight pathogen but increases susceptibility to early blight pathogen in potato via reactive oxygen species burst. N. Phytol. 2012, 196, 223–237. [Google Scholar] [CrossRef]
- Ranty, B.; Aldon, D.; Cotelle, V.; Galaud, J.P.; Thuleau, P.; Mazars, C. Calcium sensors as key hubs in plant responses to biotic and abiotic stresses. Front. Plant Sci. 2016, 7, 327. [Google Scholar] [CrossRef] [Green Version]
- Pennisi, E. Stressed Out over a Stress Hormone; American Association for the Advancement of Science: Washington, DC, USA, 2009. [Google Scholar]
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 2010, 61, 651–679. [Google Scholar] [CrossRef] [Green Version]
- Allan, A.C.; Fricker, M.D.; Ward, J.L.; Beale, M.H.; Trewavas, A.J. Two transduction pathways mediate rapid effects of abscisic acid in Commelina guard cells. Plant Cell 1994, 6, 1319–1328. [Google Scholar] [CrossRef]
- Levchenko, V.; Konrad, K.R.; Dietrich, P.; Roelfsema, M.R.G.; Hedrich, R. Cytosolic abscisic acid activates guard cell anion channels without preceding Ca2+ signals. Proc. Natl. Acad. Sci. USA 2005, 102, 4203–4208. [Google Scholar] [CrossRef] [Green Version]
- McAinsh, M.R.; Brownlee, C.; Hetherington, A.M. Abscisic acid-induced elevation of guard cell cytosolic Ca2+ precedes stomatal closure. Nature 1990, 343, 186–188. [Google Scholar] [CrossRef] [Green Version]
- Sheen, J. Ca2+-dependent protein kinases and stress signal transduction in plants. Science 1996, 274, 1900–1902. [Google Scholar] [CrossRef] [Green Version]
- MacRobbie, E.A.C. ABA activates multiple Ca2+ fluxes in stomatal guard cells, triggering vacuolar K+ (Rb+) release. Proc. Natl. Acad. Sci. USA 2000, 97, 12361–12368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Köhler, B.; Blatt, M.R. Protein phosphorylation activates the guard cell Ca2+ channel and is a prerequisite for gating by abscisic acid. Plant J. 2002, 32, 185–194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siegel, R.S.; Xue, S.; Murata, Y.; Yang, Y.; Nishimura, N.; Wang, A.; Schroeder, J.I. Calcium elevation-dependent and attenuated resting calcium-dependent abscisic acid induction of stomatal closure and abscisic acid-induced enhancement of calcium sensitivities of S-type anion and inward-rectifying K+ channels in Arabidopsis guard cells. Plant J. 2009, 59, 207–220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, H.I.; Park, H.J.; Park, J.H.; Kim, S.; Im, M.-Y.; Seo, H.H.; Kim, Y.W.; Hwang, I.; Kim, S.Y. Arabidopsis calcium-dependent protein kinase AtCPK32 interacts with ABF4, a transcriptional regulator of abscisic acid-responsive gene expression, and modulates its activity. Plant Physiol. 2005, 139, 1750–1761. [Google Scholar] [CrossRef] [Green Version]
- Mori, I.C.; Murata, Y.; Yang, Y.; Munemasa, S.; Wang, Y.F.; Andreoli, S.; Tiriac, H.; Alonso, J.M.; Harper, J.F.; Ecker, J.R. CDPKs CPK6 and CPK3 function in ABA regulation of guard cell S-type anion-and Ca2+-permeable channels and stomatal closure. PLoS Biol. 2006, 4, e327. [Google Scholar] [CrossRef] [Green Version]
- Ma, S.Y.; Wu, W.H. AtCPK23 functions in Arabidopsis responses to drought and salt stresses. Plant Mol. Biol. 2007, 65, 511–518. [Google Scholar] [CrossRef]
- Zou, J.J.; Wei, F.J.; Wang, C.; Wu, J.J.; Ratnasekera, D.; Liu, W.X.; Wu, W.H. Arabidopsis calcium-dependent protein kinase AtCPK10 functions in ABA and Ca2+-mediated stomatal regulation in response to drought stress. Plant Physiol. 2010, 154, 1232–1243. [Google Scholar] [CrossRef] [Green Version]
- Franz, S.; Ehlert, B.; Liese, A.; Kurth, J.; Cazalé, A.C.; Romeis, T. Calcium-dependent protein kinase CPK21 functions in abiotic stress response in Arabidopsis thaliana. Mol. Plant 2011, 4, 83–96. [Google Scholar] [CrossRef]
- Jiang, S.; Zhang, D.; Wang, L.; Pan, J.; Liu, Y.; Kong, X.; Zhou, Y.; Li, D. Biochemistry. A maize calcium-dependent protein kinase gene, ZmCPK4, positively regulated abscisic acid signaling and enhanced drought stress tolerance in transgenic Arabidopsis. Plant Physiol. Biochem. 2013, 71, 112–120. [Google Scholar] [CrossRef]
- Sanders, D.; Pelloux, J.; Brownlee, C.; Harper, J.F. Calcium at the crossroads of signaling. Plant Cell 2002, 14, S401–S417. [Google Scholar] [CrossRef] [Green Version]
- Wurzinger, B.; Mair, A.; Pfister, B.; Teige, M. Cross-talk of calcium-dependent protein kinase and MAP kinase signaling. Plant Signal. Behav. 2011, 6, 8–12. [Google Scholar] [CrossRef] [PubMed]
- Petersen, M.; Brodersen, P.; Naested, H.; Andreasson, E.; Lindhart, U.; Bo, J.; Nielsen, H.B.; Lacy, M.; Austin, M.J.; Parker, J.E. Arabidopsis MAP Kinase 4 Negatively Regulates Systemic Acquired Resistance. Cell 2000, 103, 1111–1120. [Google Scholar] [CrossRef] [Green Version]
- Asai, T.; Tena, G.; Plotnikova, J.; Willmann, M.R.; Chiu, W.L.; Gomez-Gomez, L.; Boller, T.; Ausubel, F.M.; Sheen, J. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 2002, 415, 977–983. [Google Scholar] [CrossRef] [PubMed]
- Mészáros, T.; Helfer, A.; Hatzimasoura, E.; Magyar, Z.; Serazetdinova, L.; Rios, G.; Bardóczy, V.; Teige, M.; Koncz, C.; Peck, S. The Arabidopsis MAP kinase kinase MKK1 participates in defence responses to the bacterial elicitor flagellin. Plant J. 2006, 48, 485–498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dóczi, R.; Brader, G.; Pettkószandtner, A.; Rajh, I.; Djamei, A.; Pitzschke, A.; Teige, M.; Hirt, H. The Arabidopsis Mitogen-Activated Protein Kinase Kinase MKK3 Is Upstream of Group C Mitogen-Activated Protein Kinases and Participates in Pathogen Signaling. Plant Cell 2007, 19, 3266–3279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agell, N.; Bachs, O.; Rocamora, N.; Villalonga, P. Modulation of the Ras/Raf/MEK/ERK pathway by Ca 2+, and Calmodulin. Cell. Signal. 2002, 14, 649–654. [Google Scholar] [CrossRef]
- Rozengurt, E. Mitogenic signaling pathways induced by G protein-coupled receptors. J. Cell. Physiol. 2007, 213, 589–602. [Google Scholar] [CrossRef]
- Boudsocq, M.; Willmann, M.; McCormack, M.; Lee, H.; Shan, L.; He, P.; Bush, J.; Cheng, S.; Sheen, J. Differential innate immune signalling via Ca2+ sensor protein kinases in plants. Nature 2010, 464, 418–422. [Google Scholar] [CrossRef] [Green Version]
- Ludwig, A.A.; Saitoh, H.; Felix, G.; Freymark, G.; Miersch, O.; Wasternack, C.; Boller, T.; Jones, J.D.G.; Romeis, T. Ethylene-mediated cross-talk between calcium-dependent protein kinase and MAPK signaling controls stress responses in plants. Proc. Natl. Acad. Sci. USA 2005, 102, 10736–10741. [Google Scholar] [CrossRef] [Green Version]
- Ding, Y.; Cao, J.; Ni, L.; Zhu, Y.; Zhang, A.; Tan, M.; Jiang, M. ZmCPK11 is involved in abscisic acid-induced antioxidant defence and functions upstream of ZmMPK5 in abscisic acid signalling in maize. J. Exp. Bot. 2013, 64, 871–884. [Google Scholar] [CrossRef] [Green Version]
- Xie, K.; Yang, Y. Direct phosphorylation and activation of a mitogen-activated protein kinase by a calcium-dependent protein kinase in rice. Plant Cell 2014, 26, 3077–3089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, P.; Wang, W.; Duan, W.; Li, Y.; Hou, X. Comprehensive Analysis of the CDPK-SnRK Superfamily Genes in Chinese Cabbage and Its Evolutionary Implications in Plants. Front. Plant Sci. 2017, 8, 162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, X.; Lv, W.; Jiang, S.; Zhang, D.; Cai, G.; Pan, J.; Li, D. Genome-wide identification and expression analysis of calcium-dependent protein kinase in maize. BMC Genom. 2013, 14, 433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boudsocq, M.; Sheen, J. CDPKs in immune and stress signaling. Trends Plant Sci. 2013, 18, 30–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morello, L.; Bardini, M.; Cricrì, M.; Sala, F.; Breviario, D. Functional analysis of DNA sequences controlling the expression of the rice OsCDPK2 gene. Planta 2006, 223, 479–491. [Google Scholar] [CrossRef] [PubMed]
- Boudsocq, M.; Droillard, M.J.; Regad, L.; Laurière, C. Characterization of Arabidopsis calcium-dependent protein kinases: Activated or not by calcium? Biochem. J. 2012, 447, 291–299. [Google Scholar] [CrossRef] [Green Version]
- Hong, Y.; Takano, M.; Liu, C.M.; Gasch, A.; Chye, M.L.; Chua, N.H. Expression of three members of the calcium-dependent protein kinase gene family in Arabidopsis thaliana. Plant Mol. Biol. 1996, 30, 1259–1275. [Google Scholar] [CrossRef] [Green Version]
- Ye, S.; Wang, L.; Xie, W.; Wan, B.; Li, X.; Lin, Y. Expression profile of calcium-dependent protein kinase (CDPKs) genes during the whole lifespan and under phytohormone treatment conditions in rice (Oryza sativa L. ssp. indica). Plant Mol. Biol. 2009, 70, 311–325. [Google Scholar] [CrossRef]
- Monaghan, J.; Matschi, S.; Shorinola, O.; Rovenich, H.; Matei, A.; Segonzac, C.; Malinovsky, F.G.; Rathjen, J.P.; Maclean, D.; Romeis, T.; et al. The calcium-dependent protein kinase CPK28 buffers plant immunity and regulates BIK1 turnover. Cell Host Microbe 2014, 16, 605–615. [Google Scholar] [CrossRef] [Green Version]
- Rudd, J.J.; Franklin Tong, V.E. Unravelling response-specificity in Ca2+ signalling pathways in plant cells. N. Phytol. 2001, 151, 7–33. [Google Scholar] [CrossRef]
- Campo, S.; Baldrich, P.; Messeguer, J.; Lalanne, E.; Coca, M.; San, S.B. Overexpression of a Calcium-Dependent Protein Kinase Confers Salt and Drought Tolerance in Rice by Preventing Membrane Lipid Peroxidation. Plant Physiol. 2014, 165, 688–704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campos-Soriano, L.; Gómez-Ariza, J.; Bonfante, P.; San Segundo, B. A rice calcium-dependent protein kinase is expressed in cortical root cells during the presymbiotic phase of the arbuscular mycorrhizal symbiosis. BMC Plant Biol. 2011, 11, 90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yun, W.; Zhang, M.; Ke, K.; Lu, Y.T. Cellular localization and biochemical characterization of a novel calcium-dependent protein kinase from tobacco. Cell Res. 2005, 15, 604–612. [Google Scholar]
- Gutermuth, T.; Lassig, R.; Portes, M.T.; Maierhofer, T.; Romeis, T.; Borst, J.W.; Hedrich, R.; Feijó, J.A.; Konrad, K.R. Pollen tube growth regulation by free anions depends on the interaction between the anion channel SLAH3 and calcium-dependent protein kinases CPK2 and CPK20. Plant Cell 2013, 25, 4525–4543. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Che, Z.; Zeng, X.; Zhou, X.; Sitoe, H.M.; Wang, H.; Yu, D.J.F.; Genomics, I. Genome-wide analysis of calcium-dependent protein kinases and their expression patterns in response to herbivore and wounding stresses in soybean. Funct. Integr. Genom. 2016, 16, 481–493. [Google Scholar] [CrossRef]
- Curran, A.; Chang, I.F.; Chang, C.L.; Garg, S.; Miguel, R.M.; Barron, Y.D.; Li, Y.; Romanowsky, S.; Cushman, J.C.; Gribskov, M. Calcium-dependent protein kinases from Arabidopsis show substrate specificity differences in an analysis of 103 substrates. Front. Plant Sci. 2011, 2, 36. [Google Scholar] [CrossRef] [Green Version]
- Klimecka, M.; Muszynska, G. Structure and functions of plant calcium-dependent protein kinases. Acta Biochim. Pol. Engl. Ed. 2007, 54, 219–233. [Google Scholar] [CrossRef] [Green Version]
- Soon, F.F.; Ng, L.M.; Zhou, X.E.; West, G.M.; Kovach, A.; Tan, M.E.; Suino-Powell, K.M.; He, Y.; Xu, Y.; Chalmers, M.J.; et al. Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases. Science 2012, 335, 85–88. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.G.; Fu, F.L.; Yu, H.Q.; Hu, T.; Zhang, Y.Y.; Tao, Y.; Zhu, J.K.; Zhao, Y.; Li, W.C. Interaction network of core ABA signaling components in maize. Plant Mol. Biol. 2018, 96, 245–263. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, W.Z.; Zhang, Y.; Deng, M.; Niu, F.; Yang, B.; Wang, X.; Wang, B.; Liang, W.; Deyholos, M.K.; et al. Identification, expression and interaction analyses of calcium-dependent protein kinase (CPK) genes in canola (Brassica napus L.). BMC Genom. 2014, 15, 211. [Google Scholar] [CrossRef] [Green Version]
- Lynch, T.; Erickson, B.J.; Finkelstein, R.R. Direct interactions of ABA-insensitive (ABI)-clade protein phosphatase (PP) 2Cs with calcium-dependent protein kinases and ABA response element-binding bZIPs may contribute to turning off ABA response. Plant Mol. Biol. 2012, 80, 647–658. [Google Scholar] [CrossRef] [PubMed]
- Belin, C.; De Franco, P.O.; Bourbousse, C.; Chaignepain, S.; Schmitter, J.M.; Vavasseur, A.; Giraudat, J.; Barbier-Brygoo, H.; Thomine, S. Identification of features regulating OST1 kinase activity and OST1 function in guard cells. Plant Physiol. 2006, 141, 1316–1327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshida, R.; Umezawa, T.; Mizoguchi, T.; Takahashi, S.; Takahashi, F.; Shinozaki, K. The regulatory domain of SRK2E/OST1/SnRK2. 6 interacts with ABI1 and integrates abscisic acid (ABA) and osmotic stress signals controlling stomatal closure in Arabidopsis. J. Biol. Chem. 2006, 281, 5310–5318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshida, T.; Nishimura, N.; Kitahata, N.; Kuromori, T.; Ito, T.; Asami, T.; Shinozaki, K.; Hirayama, T. ABA-hypersensitive germination3 encodes a protein phosphatase 2C (AtPP2CA) that strongly regulates abscisic acid signaling during germination among Arabidopsis protein phosphatase 2Cs. Plant Physiol. 2006, 140, 115–126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mustilli, A.C.; Merlot, S.; Vavasseur, A.; Fenzi, F.; Giraudat, J. Arabidopsis OST1 Protein Kinase Mediates the Regulation of Stomatal Aperture by Abscisic Acid and Acts Upstream of Reactive Oxygen Species Production. Plant Cell 2002, 14, 3089–3099. [Google Scholar] [CrossRef] [Green Version]
- Vlad, F.; Rubio, S.; Rodrigues, A.; Sirichandra, C.; Belin, C.; Robert, N.; Leung, J.; Rodriguez, P.L.; Laurière, C.; Merlot, S. Protein Phosphatases 2C Regulate the Activation of the Snf1-Related Kinase OST1 by Abscisic Acid in Arabidopsis. Plant Cell 2009, 21, 3170–3184. [Google Scholar] [CrossRef] [Green Version]
- Kulik, A.; Wawer, I.; Krzywińska, E.; Bucholc, M.; Dobrowolska, G. SnRK2 Protein Kinases—Key Regulators of Plant Response to Abiotic Stresses. OMICS J. Integr. Biol. 2011, 15, 859–872. [Google Scholar] [CrossRef]
- Xu, C.; Li, X.; Zhang, L. The effect of calcium chloride on growth, photosynthesis, and antioxidant responses of Zoysia japonica under drought conditions. PLoS ONE 2013, 8, e68214. [Google Scholar] [CrossRef]
- Yang, C.; Liu, J.; Dong, X.; Cai, Z.; Tian, W.; Wang, X. Short-term and continuing stresses differentially interplay with multiple hormones to regulate plant survival and growth. Mol. Plant 2014, 7, 841–855. [Google Scholar] [CrossRef] [Green Version]
- Patykowski, J.; Kołodziejek, J.; Wala, M. Biochemical and growth responses of silver maple (Acer saccharinum L.) to sodium chloride and calcium chloride. PeerJ 2018, 6, e5958. [Google Scholar] [CrossRef] [Green Version]
- Lin, F.; Jiang, L.; Liu, Y.; Lv, Y.; Dai, H.; Zhao, H. Genome-wide identification of housekeeping genes in maize. Plant Mol. Biol. 2014, 86, 543–554. [Google Scholar] [CrossRef] [PubMed]







© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khalid, M.H.B.; Raza, M.A.; Yu, H.Q.; Khan, I.; Sun, F.A.; Feng, L.Y.; Qu, J.T.; Fu, F.L.; Li, W.C. Expression, Subcellular Localization, and Interactions of CPK Family Genes in Maize. Int. J. Mol. Sci. 2019, 20, 6173. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20246173
Khalid MHB, Raza MA, Yu HQ, Khan I, Sun FA, Feng LY, Qu JT, Fu FL, Li WC. Expression, Subcellular Localization, and Interactions of CPK Family Genes in Maize. International Journal of Molecular Sciences. 2019; 20(24):6173. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20246173
Chicago/Turabian StyleKhalid, Muhammad Hayder Bin, Muhammad Ali Raza, Hao Qiang Yu, Imran Khan, Fu Ai Sun, Ling Yang Feng, Jing Tao Qu, Feng Ling Fu, and Wan Chen Li. 2019. "Expression, Subcellular Localization, and Interactions of CPK Family Genes in Maize" International Journal of Molecular Sciences 20, no. 24: 6173. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20246173