GeneLab Database Analyses Suggest Long-Term Impact of Space Radiation on the Cardiovascular System by the Activation of FYN Through Reactive Oxygen Species
Abstract
:1. Introduction
2. Results
2.1. Global Analysis of Simulated Space Radiation Ground Experiments Compared to Spaceflight Samples
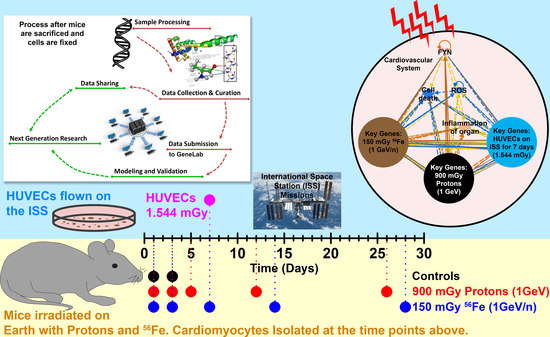
2.2. Dose Received by HUVECs Flown in Space
2.3. Common Pathways between Simulated Space Radiation and Spaceflight Samples
2.4. The Biological Pathways Elicited by Protons Have Stronger Overlap with Spaceflight Samples Than Those Elicited by 56Fe Ions
2.5. Key Driving Genes Provide Direct Connection to ROS for Space Radiation Impact on the Cardiovascular System
2.6. Predicted Circulating miRNA Signature Associated with Space Radiation Cardiovascular Risk
3. Discussion
4. Materials and Methods
4.1. Datasets Utilized from the GeneLab Platform
4.2. Transcriptomic Analysis
4.3. Determination of Key Genes/Drivers
4.4. MicroRNA (miRNA) Predictions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ESA-SPHINX | European Space Agency-Integrated Experiment |
| SLPSRA | Space Life and Physical Sciences Research and Applications |
| HUVECs | Human Umbilical Vein Endothelial Cells |
| ANOVA | Analysis of Variance |
| miRNA | microRNA |
| GLDS | GeneLab Dataset |
| GSEA | Gene Set Enrichment Analysis |
| NASA | National Aeronautics and Space Administration |
| NSRL | NASA Space Radiation Laboratory |
| BNL | Brookhaven National Laboratory |
| CNS | Central Nervous System |
| ESA | European Space Agency |
| FDR | False Discovery Rate |
| GCR | Galactic Cosmic Rays |
| HZE | High (H) Atomic number (Z) and Energy (E) |
| 56Fe | Iron |
| IPA | Ingenuity Pathway Analysis |
| ISS | International Space Station |
| LEO | Low Earth Orbit |
| LET | Linear Energy Transfer |
| MeV | Mega Electron Volt |
| PCA | Principal Component Analysis |
| ROS | Reactive Oxygen Species |
| SAA | South Atlantic Anomaly |
| SPE | Solar Particle Event |
| Gy | Gray |
| PC | Principal Component |
References
- Boerma, M.; Nelson, G.A.; Sridharan, V.; Mao, X.W.; Koturbash, I.; Hauer-Jensen, M. Space radiation and cardiovascular disease risk. World J. Cardiol. 2015, 7, 882–888. [Google Scholar] [CrossRef] [PubMed]
- White, R.J.; Averner, M. Humans in space. Nature 2001, 409, 1115–1118. [Google Scholar] [CrossRef] [PubMed]
- Durante, M.; Cucinotta, F.A. Heavy ion carcinogenesis and human space exploration. Nat. Rev. Cancer 2008, 8, 465–472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, T.; Chen, H.; Lv, K.; Ji, G.; Liang, F.; Zhang, Y.; Wang, Y.; Liu, X.; Cao, H.; Kan, G.; et al. Activation of HIF-1alpha and its downstream targets in rat hippocampus after long-term simulated microgravity exposure. Biochem. Biophys. Res. Commun 2017, 485, 591–597. [Google Scholar] [CrossRef] [PubMed]
- Barcellos-Hoff, M.H.; Mao, J.H. HZE radiation non-targeted effects on the microenvironment that mediate mammary carcinogenesis. Front. Oncol. 2016, 6, 57. [Google Scholar] [CrossRef] [PubMed]
- Cucinotta, F.A.; Schimmerling, W.; Wilson, J.W.; Peterson, L.E.; Badhwar, G.D.; Saganti, P.B.; Dicello, J.F. Space radiation cancer risks and uncertainties for Mars missions. Radiat. Res. 2001, 156, 682–688. [Google Scholar] [CrossRef]
- Grabham, P.; Hu, B.; Sharma, P.; Geard, C. Effects of ionizing radiation on three-dimensional human vessel models: Differential effects according to radiation quality and cellular development. Radiat. Res. 2011, 175, 21–28. [Google Scholar] [CrossRef]
- Jain, M.R.; Li, M.; Chen, W.; Liu, T.; de Toledo, S.M.; Pandey, B.N.; Li, H.; Rabin, B.M.; Azzam, E.I. In vivo space radiation-induced non-targeted responses: Late effects on molecular signaling in mitochondria. Curr. Mol. Pharmacol. 2011, 4, 106–114. [Google Scholar] [CrossRef]
- Moreno-Villanueva, M.; Wong, M.; Lu, T.; Zhang, Y.; Wu, H. Interplay of space radiation and microgravity in DNA damage and DNA damage response. NPJ Microgravity 2017, 3, 14. [Google Scholar] [CrossRef]
- Cortese, F.; Klokov, D.; Osipov, A.; Stefaniak, J.; Moskalev, A.; Schastnaya, J.; Cantor, C.; Aliper, A.; Mamoshina, P.; Ushakov, I.; et al. Vive la radioresistance!: Converging research in radiobiology and biogerontology to enhance human radioresistance for deep space exploration and colonization. Oncotarget 2018, 9, 14692–14722. [Google Scholar] [CrossRef]
- Beheshti, A.; Peluso, M.; Lamont, C.; Hahnfeldt, P.; Hlatky, L. Proton irradiation augments the suppression of tumor progression observed with advanced age. Radiat. Res. 2014, 181, 272–283. [Google Scholar] [CrossRef] [PubMed]
- Beheshti, A.; Sachs, R.K.; Peluso, M.; Rietman, E.; Hahnfeldt, P.; Hlatky, L. Age and space irradiation modulate tumor progression: Implications for carcinogenesis risk. Radiat. Res. 2013, 179, 208–220. [Google Scholar] [CrossRef] [PubMed]
- Chancellor, J.C.; Scott, G.B.; Sutton, J.P. Space radiation: The number one risk to astronaut health beyond low earth orbit. Life (Basel) 2014, 4, 491–510. [Google Scholar] [CrossRef] [PubMed]
- Alpen, E.L.; Powers-Risius, P.; Curtis, S.B.; DeGuzman, R.; Fry, R.J. Fluence-based relative biological effectiveness for charged particle carcinogenesis in mouse harderian gland. Adv. Space Res. 1994, 14, 573–581. [Google Scholar] [CrossRef]
- Cucinotta, F.A.; To, K.; Cacao, E. Predictions of space radiation fatality risk for exploration missions. Life Sci. Space Res. 2017, 13, 1–11. [Google Scholar] [CrossRef]
- Townsend, L.W.; Adams, J.H.; Blattnig, S.R.; Clowdsley, M.S.; Fry, D.J.; Jun, I.; McLeod, C.D.; Minow, J.I.; Moore, D.F.; Norbury, J.W.; et al. Solar particle event storm shelter requirements for missions beyond low earth orbit. Life Sci. Space Res. 2018, 17, 32–39. [Google Scholar] [CrossRef]
- Beheshti, A.; Miller, J.; Kidane, Y.; Berrios, D.; Gebre, S.G.; Costes, S.V. Nasa GeneLab project: Bridging space radiation omics with ground studies. Radiat. Res. 2018, 189, 553–559. [Google Scholar] [CrossRef]
- Gridley, D.S.; Obenaus, A.; Bateman, T.A.; Pecaut, M.J. Long-term changes in rat hematopoietic and other physiological systems after high-energy iron ion irradiation. Int. J. Radiat. Biol 2008, 84, 549–559. [Google Scholar] [CrossRef]
- Park, J.S.; Qiao, L.; Su, Z.Z.; Hinman, D.; Willoughby, K.; McKinstry, R.; Yacoub, A.; Duigou, G.J.; Young, C.S.; Grant, S.; et al. Ionizing radiation modulates vascular endothelial growth factor (VEGF) expression through multiple mitogen activated protein kinase dependent pathways. Oncogene 2001, 20, 3266–3280. [Google Scholar] [CrossRef] [Green Version]
- Hughson, R.L.; Helm, A.; Durante, M. Heart in space: Effect of the extraterrestrial environment on the cardiovascular system. Nat. Rev. Cardiol. 2018, 15, 167–180. [Google Scholar] [CrossRef]
- Little, M.P.; Azizova, T.V.; Bazyka, D.; Bouffler, S.D.; Cardis, E.; Chekin, S.; Chumak, V.V.; Cucinotta, F.A.; de Vathaire, F.; Hall, P.; et al. Systematic review and meta-analysis of circulatory disease from exposure to low-level ionizing radiation and estimates of potential population mortality risks. Environ. Health Perspect. 2012, 120, 1503–1511. [Google Scholar] [CrossRef] [PubMed]
- Yamada, M.; Wong, F.L.; Fujiwara, S.; Akahoshi, M.; Suzuki, G. Noncancer disease incidence in atomic bomb survivors, 1958–1998. Radiat. Res. 2004, 161, 622–632. [Google Scholar] [CrossRef] [PubMed]
- Stewart, A.M.; Kneale, G.W. A-bomb survivors: Factors that may lead to a re-assessment of the radiation hazard. Int. J. Epidemiol. 2000, 29, 708–714. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Sasi, S.P.; Gee, H.; Lee, J.; Yang, Y.; Mehrzad, R.; Onufrak, J.; Song, J.; Enderling, H.; Agarwal, A.; et al. Cardiovascular risks associated with low dose ionizing particle radiation. PLoS ONE 2014, 9, e110269. [Google Scholar] [CrossRef] [PubMed]
- Tungjai, M.; Whorton, E.B.; Rithidech, K.N. Persistence of apoptosis and inflammatory responses in the heart and bone marrow of mice following whole-body exposure to 28silicon (28Si) ions. Radiat. Environ. Biophys. 2013, 52, 339–350. [Google Scholar] [CrossRef] [PubMed]
- Lima, F.; Ding, D.; Goetz, W.; Yang, A.J.; Baulch, J.E. High LET 56Fe ion irradiation induces tissue-specific changes in DNA methylation in the mouse. Environ. Mol. Mutagen. 2014, 55, 266–277. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Parks, B.W.; Yu, S.; Srivastava, R.; Gupta, K.; Wu, X.; Khaled, S.; Chang, P.Y.; Kabarowski, J.H.; Kucik, D.F. Iron-ion radiation accelerates atherosclerosis in apolipoprotein E-deficient mice. Radiat. Res. 2011, 175, 766–773. [Google Scholar] [CrossRef]
- Grabham, P.; Sharma, P.; Bigelow, A.; Geard, C. Two distinct types of the inhibition of vasculogenesis by different species of charged particles. Vasc. Cell 2013, 5, 16. [Google Scholar] [CrossRef] [Green Version]
- Koturbash, I.; Miousse, I.R.; Sridharan, V.; Nzabarushimana, E.; Skinner, C.M.; Melnyk, S.B.; Pavliv, O.; Hauer-Jensen, M.; Nelson, G.A.; Boerma, M. Radiation-induced changes in DNA methylation of repetitive elements in the mouse heart. Mutat. Res. 2016, 787, 43–53. [Google Scholar] [CrossRef] [Green Version]
- Soucy, K.G.; Lim, H.K.; Kim, J.H.; Oh, Y.; Attarzadeh, D.O.; Sevinc, B.; Kuo, M.M.; Shoukas, A.A.; Vazquez, M.E.; Berkowitz, D.E. HZE 56Fe-ion irradiation induces endothelial dysfunction in rat aorta: Role of xanthine oxidase. Radiat. Res. 2011, 176, 474–485. [Google Scholar] [CrossRef]
- Kim, D.J.; Norden, P.R.; Salvador, J.; Barry, D.M.; Bowers, S.L.K.; Cleaver, O.; Davis, G.E. Src- and FYN-dependent apical membrane trafficking events control endothelial lumen formation during vascular tube morphogenesis. PLoS ONE 2017, 12, e0184461. [Google Scholar] [CrossRef] [PubMed]
- Beneduce, E.; Matte, A.; De Falco, L.; Mbiandjeu, S.; Chiabrando, D.; Tolosano, E.; Federti, E.; Petrillo, S.; Mohandas, N.; Siciliano, A.; et al. Fyn kinase is a novel modulator of erythropoietin signaling and stress erythropoiesis. Am. J. Hematol. 2018, 94, 10–20. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, S.M.; Lemmens-Gruber, R. Phosphorylation of cardiac voltage-gated sodium channel: Potential players with multiple dimensions. Acta Physiol. 2018, e13210. [Google Scholar] [CrossRef]
- Unoki, T.; Akiyama, M.; Kumagai, Y.; Goncalves, F.M.; Farina, M.; da Rocha, J.B.T.; Aschner, M. Molecular pathways associated with methylmercury-induced NRF2 modulation. Front. Genet. 2018, 9, 373. [Google Scholar] [CrossRef] [PubMed]
- Matsushima, S.; Kuroda, J.; Zhai, P.; Liu, T.; Ikeda, S.; Nagarajan, N.; Oka, S.; Yokota, T.; Kinugawa, S.; Hsu, C.P.; et al. Tyrosine kinase FYN negatively regulates NOX4 in cardiac remodeling. J. Clin. Investig. 2016, 126, 3403–3416. [Google Scholar] [CrossRef] [Green Version]
- Palacios, E.H.; Weiss, A. Function of the SRC-family kinases, LCK and FYN, in T-cell development and activation. Oncogene 2004, 23, 7990–8000. [Google Scholar] [CrossRef] [PubMed]
- Abe, J.; Berk, B.C. FYN and JAK2 mediate RAS activation by reactive oxygen species. J. Biol. Chem. 1999, 274, 21003–21010. [Google Scholar] [CrossRef] [PubMed]
- Qin, Q.; Qu, C.; Niu, T.; Zang, H.; Qi, L.; Lyu, L.; Wang, X.; Nagarkatti, M.; Nagarkatti, P.; Janicki, J.S.; et al. NRF2-mediated cardiac maladaptive remodeling and dysfunction in a setting of autophagy insufficiency. Hypertension 2016, 67, 107–117. [Google Scholar] [CrossRef] [PubMed]
- Saito, Y.D.; Jensen, A.R.; Salgia, R.; Posadas, E.M. FYN: A novel molecular target in cancer. Cancer 2010, 116, 1629–1637. [Google Scholar] [CrossRef] [PubMed]
- Elias, D.; Ditzel, H.J. Fyn is an important molecule in cancer pathogenesis and drug resistance. Pharmacol. Res. 2015, 100, 250–254. [Google Scholar] [CrossRef] [PubMed]
- Mkaddem, S.B.; Murua, A.; Flament, H.; Titeca-Beauport, D.; Bounaix, C.; Danelli, L.; Launay, P.; Benhamou, M.; Blank, U.; Daugas, E.; et al. LYN and FYN function as molecular switches that control immunoreceptors to direct homeostasis or inflammation. Nat. Commun. 2017, 8, 246. [Google Scholar] [CrossRef] [PubMed]
- Gonsalvez, D.; Ferner, A.H.; Peckham, H.; Murray, S.S.; Xiao, J. The roles of extracellular related-kinases 1 and 2 signaling in CNS myelination. Neuropharmacology 2016, 110, 586–593. [Google Scholar] [CrossRef] [PubMed]
- Versari, S.; Longinotti, G.; Barenghi, L.; Maier, J.A.; Bradamante, S. The challenging environment on board the international space station affects endothelial cell function by triggering oxidative stress through thioredoxin interacting protein overexpression: The ESA-SPHINX experiment. FASEB J. 2013, 27, 4466–4475. [Google Scholar] [CrossRef] [PubMed]
- Coleman, M.A.; Sasi, S.P.; Onufrak, J.; Natarajan, M.; Manickam, K.; Schwab, J.; Muralidharan, S.; Peterson, L.E.; Alekseyev, Y.O.; Yan, X.; et al. Low-dose radiation affects cardiac physiology: Gene networks and molecular signaling in cardiomyocytes. Am. J. Physiol. Heart Circ. Physiol. 2015, 309, H1947–H1963. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quijano, C.; Trujillo, M.; Castro, L.; Trostchansky, A. Interplay between oxidant species and energy metabolism. Redox Biol. 2016, 8, 28–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.R.; Zweier, J.L. Cardiac mitochondria and reactive oxygen species generation. Circ. Res. 2014, 114, 524–537. [Google Scholar] [CrossRef]
- Edmunds, L.R.; Sharma, L.; Kang, A.; Lu, J.; Vockley, J.; Basu, S.; Uppala, R.; Goetzman, E.S.; Beck, M.E.; Scott, D.; et al. C-MYC programs fatty acid metabolism and dictates acetyl-coA abundance and fate. J. Biol. Chem. 2014, 289, 25382–25392. [Google Scholar] [CrossRef]
- Kanaan, G.N.; Harper, M.E. Cellular redox dysfunction in the development of cardiovascular diseases. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 2822–2829. [Google Scholar] [CrossRef]
- de Villiers, D.; Potgieter, M.; Ambele, M.A.; Adam, L.; Durandt, C.; Pepper, M.S. The role of reactive oxygen species in adipogenic differentiation. Adv. Exp. Med. Biol. 2017, 1083, 125–144. [Google Scholar] [CrossRef]
- He, F.; Zuo, L. Redox roles of reactive oxygen species in cardiovascular diseases. Int J. Mol. Sci. 2015, 16, 27770–27780. [Google Scholar] [CrossRef] [PubMed]
- Verbon, E.H.; Post, J.A.; Boonstra, J. The influence of reactive oxygen species on cell cycle progression in mammalian cells. Gene 2012, 511, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Beheshti, A.; Ray, S.; Fogle, H.; Berrios, D.; Costes, S.V. A microRNA signature and TGF-beta1 response were identified as the key master regulators for spaceflight response. PLoS ONE 2018, 13, e0199621. [Google Scholar] [CrossRef] [PubMed]
- Wage, J.; Ma, L.; Peluso, M.; Lamont, C.; Evens, A.M.; Hahnfeldt, P.; Hlatky, L.; Beheshti, A. Proton irradiation impacts age-driven modulations of cancer progression influenced by immune system transcriptome modifications from splenic tissue. J. Radiat. Res. 2015, 56, 792–803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beheshti, A.; Benzekry, S.; McDonald, J.T.; Ma, L.; Peluso, M.; Hahnfeldt, P.; Hlatky, L. Host age is a systemic regulator of gene expression impacting cancer progression. Cancer Res. 2015, 75, 1134–1143. [Google Scholar] [CrossRef] [PubMed]
- Beheshti, A.; Cekanaviciute, E.; Smith, D.J.; Costes, S.V. Global transcriptomic analysis suggests carbon dioxide as an environmental stressor in spaceflight: A systems biology genelab case study. Sci. Rep. 2018, 8, 4191. [Google Scholar] [CrossRef] [PubMed]
- Beheshti, A.; Neuberg, D.; McDonald, J.T.; Vanderburg, C.R.; Evens, A.M. The impact of age and sex in dlbcl: Systems biology analyses identify distinct molecular changes and signaling networks. Cancer Inform. 2015, 14, 141–148. [Google Scholar] [CrossRef]
- Beheshti, A.; Wage, J.; McDonald, J.T.; Lamont, C.; Peluso, M.; Hahnfeldt, P.; Hlatky, L. Tumor-host signaling interaction reveals a systemic, age-dependent splenic immune influence on tumor development. Oncotarget 2015, 6, 35419–35432. [Google Scholar] [CrossRef]
- Ravi, D.; Beheshti, A.; Abermil, N.; Passero, F.; Sharma, J.; Coyle, M.; Kritharis, A.; Kandela, I.; Hlatky, L.; Sitkovsky, M.V.; et al. Proteasomal inhibition by ixazomib induces CHK1 and MYC-dependent cell death in T-cell and hodgkin lymphoma. Cancer Res. 2016, 76, 3319–3331. [Google Scholar] [CrossRef]
- Lannutti, B.J.; Drachman, J.G. Lyn tyrosine kinase regulates thrombopoietin-induced proliferation of hematopoietic cell lines and primary megakaryocytic progenitors. Blood 2004, 103, 3736–3743. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.G.; Aikawa, M. Toll-like receptors and src-family kinases in atherosclerosis—Focus on macrophages. Circ. J. 2015, 79, 2332–2334. [Google Scholar] [CrossRef]
- Han, J.; Zhang, G.; Welch, E.J.; Liang, Y.; Fu, J.; Vogel, S.M.; Lowell, C.A.; Du, X.; Cheresh, D.A.; Malik, A.B.; et al. A critical role for lyn kinase in strengthening endothelial integrity and barrier function. Blood 2013, 122, 4140–4149. [Google Scholar] [CrossRef] [PubMed]
- Lowenberg, M.; Verhaar, A.P.; Bilderbeek, J.; Marle, J.; Buttgereit, F.; Peppelenbosch, M.P.; van Deventer, S.J.; Hommes, D.W. Glucocorticoids cause rapid dissociation of a T-cell-receptor-associated protein complex containing LCK and FYN. EMBO Rep. 2006, 7, 1023–1029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsia, C.H.; Jayakumar, T.; Sheu, J.R.; Tsao, S.Y.; Velusamy, M.; Hsia, C.W.; Chou, D.S.; Chang, C.C.; Chung, C.L.; Khamrang, T.; et al. Structure-antiplatelet activity relationships of novel ruthenium (ii) complexes: Investigation of its molecular targets. Molecules 2018, 23, 477. [Google Scholar] [CrossRef] [PubMed]
- Jayakumar, T.; Hsu, C.Y.; Khamrang, T.; Hsia, C.H.; Hsia, C.W.; Manubolu, M.; Sheu, J.R. Possible molecular targets of novel ruthenium complexes in antiplatelet therapy. Int. J. Mol. Sci. 2018, 19, 1818. [Google Scholar] [CrossRef]
- Harrison, M.J.; Chimen, M.; Hussain, M.; Iqbal, A.J.; Senis, Y.A.; Nash, G.B.; Watson, S.P.; Rainger, G.E. Signalling through SRC family kinase isoforms is not redundant in models of thrombo-inflammatory vascular disease. J. Cell. Mol. Med. 2018, 22, 4317–4327. [Google Scholar] [CrossRef]
- Dai, X.; Yan, X.; Zeng, J.; Chen, J.; Wang, Y.; Chen, J.; Li, Y.; Barati, M.T.; Wintergerst, K.A.; Pan, K.; et al. Elevating CXCR7 improves angiogenic function of EPCs via AKT/GSK-3beta/FYN-mediated NRF2 activation in diabetic limb ischemia. Circ. Res. 2017, 120, e7–e23. [Google Scholar] [CrossRef]
- Rizvi, F.; Shukla, S.; Kakkar, P. Essential role of PH domain and leucine-rich repeat protein phosphatase 2 in NRF2 suppression via modulation of AKT/GSK3beta/FYN kinase axis during oxidative hepatocellular toxicity. Cell Death Dis. 2014, 5, e1153. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pages, F.; Trajanoski, Z.; Galon, J. Cluego: A cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef]
- Condorelli, G.; Latronico, M.V.; Dorn, G.W., 2nd. MicroRNAs in heart disease: Putative novel therapeutic targets? Eur. Heart J. 2010, 31, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Dong, Y.H.; Du, W.; Shi, C.Y.; Wang, K.; Tariq, M.A.; Wang, J.X.; Li, P.F. The role of micrornas in myocardial infarction: From molecular mechanism to clinical application. Int. J. Mol. Sci. 2017, 18, 745. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q. Role of NRF2 in oxidative stress and toxicity. Annu. Rev. Pharmacol. Toxicol. 2013, 53, 401–426. [Google Scholar] [CrossRef] [PubMed]
- Vomund, S.; Schafer, A.; Parnham, M.J.; Brune, B.; von Knethen, A. Nrf2, the master regulator of anti-oxidative responses. Int. J. Mol. Sci. 2017, 18, 2772. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Okumura, H.; Guo, R.; Naruse, K. Effect of oxidative stress on cardiovascular system in response to gravity. Int. J. Mol. Sci. 2017, 18, 1426. [Google Scholar] [CrossRef] [PubMed]
- Kamo, T.; Akazawa, H.; Komuro, I. Cardiac nonmyocytes in the hub of cardiac hypertrophy. Circ. Res. 2015, 117, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Azzam, E.I.; Jay-Gerin, J.P.; Pain, D. Ionizing radiation-induced metabolic oxidative stress and prolonged cell injury. Cancer Lett. 2012, 327, 48–60. [Google Scholar] [CrossRef] [Green Version]
- Siti, H.N.; Kamisah, Y.; Kamsiah, J. The role of oxidative stress, antioxidants and vascular inflammation in cardiovascular disease (a review). Vasc. Pharmacol. 2015, 71, 40–56. [Google Scholar] [CrossRef]
- Bolstad, B.M.; Irizarry, R.A.; Astrand, M.; Speed, T.P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 2003, 19, 185–193. [Google Scholar] [CrossRef] [Green Version]
- Reich, M.; Liefeld, T.; Gould, J.; Lerner, J.; Tamayo, P.; Mesirov, J.P. Genepattern 2.0. Nat. Genet. 2006, 38, 500–501. [Google Scholar] [CrossRef]
- Saeed, A.I.; Bhagabati, N.K.; Braisted, J.C.; Liang, W.; Sharov, V.; Howe, E.A.; Li, J.; Thiagarajan, M.; White, J.A.; Quackenbush, J. TM4 microarray software suite. Methods Enzymol. 2006, 411, 134–193. [Google Scholar] [PubMed]
- Dalman, M.R.; Deeter, A.; Nimishakavi, G.; Duan, Z.H. Fold change and p-value cutoffs significantly alter microarray interpretations. BMC Bioinform. 2012, 13 (Suppl. 2), S11. [Google Scholar] [CrossRef] [Green Version]
- Paley, S.; Parker, K.; Spaulding, A.; Tomb, J.F.; O’Maille, P.; Karp, P.D. The omics dashboard for interactive exploration of gene-expression data. Nucleic Acids Res. 2017, 45, 12113–12124. [Google Scholar] [CrossRef] [PubMed]
- Kramer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef] [PubMed]







© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Beheshti, A.; McDonald, J.T.; Miller, J.; Grabham, P.; Costes, S.V. GeneLab Database Analyses Suggest Long-Term Impact of Space Radiation on the Cardiovascular System by the Activation of FYN Through Reactive Oxygen Species. Int. J. Mol. Sci. 2019, 20, 661. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20030661
Beheshti A, McDonald JT, Miller J, Grabham P, Costes SV. GeneLab Database Analyses Suggest Long-Term Impact of Space Radiation on the Cardiovascular System by the Activation of FYN Through Reactive Oxygen Species. International Journal of Molecular Sciences. 2019; 20(3):661. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20030661
Chicago/Turabian StyleBeheshti, Afshin, J. Tyson McDonald, Jack Miller, Peter Grabham, and Sylvain V. Costes. 2019. "GeneLab Database Analyses Suggest Long-Term Impact of Space Radiation on the Cardiovascular System by the Activation of FYN Through Reactive Oxygen Species" International Journal of Molecular Sciences 20, no. 3: 661. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20030661