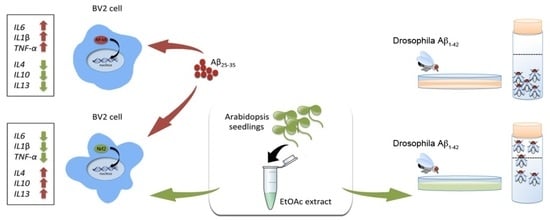
Anti-Inflammatory Activity of A Polyphenolic Extract from Arabidopsis thaliana in In Vitro and In Vivo Models of Alzheimer’s Disease
Abstract
:1. Introduction
2. Results
2.1. Qualitative and Quantitative Analysis of Polyphenols in Arabidopsis Extract
2.2. The Arabidopsis EtOAc Extract Affects Pro-Inflammatory and Anti-Inflammatory Cytokines Gene Expression in BV2 Cells
2.3. The Arabidopsis EtOAc Extract Affects Nrf2 and p65 Nuclear Translocation in BV2 Cells
2.4. The Arabidopsis EtOAc Extract Affects Cellular Antioxidant Defense Capacity and Counteracts Aβ25–35 Induced Cytotoxicity
2.5. The Arabidopsis EtOAC Extract Alleviates the Locomotor Dysfunction Induced by Aβ1–42 in Drosophila melanogaster
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Plant Growth and Juice Preparation
4.3. Determination of Total Phenols and Anthocyanins
4.4. Chromatography and Mass Spectrometry
4.5. DPPH Free Radical Scavenging Assay
4.6. Preparation of Aβ25–35 Stock Solution
4.7. Cell Culture and Treatment
4.8. Cell Viability Assay
4.9. qRT-PCR
4.10. NAD(P)H:Quinone Oxidoreductase 1 (NQO1) Activity
4.11. Nrf2 and NF-κB Immunofluorescence
4.12. Fly Stocks and Crosses
4.13. Climbing Assay
4.14. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| Aβ | Amyloid beta peptide |
| AD | Alzheimer’s disease |
| DPPH | 2,2-Diphenyl-1-picrylhydrazyl |
| EtOAc | Ethyl acetate |
| HO-1 | Heme Oxygenase 1 |
| NFκB | Nuclear factor kappa-light-chain-enhancer of activated B cells |
| NQO1 | NAD(P)H:Quinone oxidoreductase 1 |
| Nrf2 | Nuclear factor E2-related factor 2 |
| PBS | Phosphate buffer saline |
| ROS | Reactive oxygen species |
References
- Scalbert, A.; Manach, C.; Morand, C.; Remesy, C.; Jimenez, L. Dietary polyphenols and the prevention of diseases. Crit. Rev. Food Sci. Nutr. 2005, 45, 287–306. [Google Scholar] [CrossRef]
- Graf, B.A.; Milbury, P.E.; Blumberg, J.B. Flavonols, flavones, flavanones, and human health: Epidemiological evidence. J. Med. Food 2005, 8, 281–290. [Google Scholar] [CrossRef] [PubMed]
- Arts, I.C.W.; Hollman, P.C.H. Polyphenols and disease risk in epidemiologic studies. Am. J. Clin. Nutr. 2005, 81, 317s–325s. [Google Scholar] [CrossRef] [PubMed]
- Pandey, K.B.; Rizvi, S.I. Plant polyphenols as dietary antioxidants in human health and disease. Oxid. Med. Cell. Longev. 2009, 2, 270–278. [Google Scholar] [CrossRef]
- Ferrer, I. Defining Alzheimer as a common age-related neurodegenerative process not inevitably leading to dementia. Prog. Neurobiol. 2012, 97, 38–51. [Google Scholar] [CrossRef] [PubMed]
- Martire, S.; Fuso, A.; Rotili, D.; Tempera, I.; Giordano, C.; De Zottis, I.; Muzi, A.; Vernole, P.; Graziani, G.; Lococo, E.; et al. PARP-1 Modulates Amyloid Beta Peptide-Induced Neuronal Damage. PLoS ONE 2013, 8, e72169. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.C.; Zhong, C.J. Oxidative stress in Alzheimer’s disease. Neurosci. Bull. 2014, 30, 271–281. [Google Scholar] [CrossRef] [Green Version]
- Darvesh, A.S.; Carroll, R.T.; Bishayee, A.; Geldenhuys, W.J.; Van der Schyf, C.J. Oxidative stress and Alzheimer’s disease: Dietary polyphenols as potential therapeutic agents. Expert Rev. Neurother. 2010, 10, 729–745. [Google Scholar] [CrossRef]
- Jayasena, T.; Poljak, A.; Smythe, G.; Braidy, N.; Munch, G.; Sachdev, P. The role of polyphenols in the modulation of sirtuins and other pathways involved in Alzheimer’s disease. Ageing Res. Rev. 2013, 12, 867–883. [Google Scholar] [CrossRef]
- Rong, H.; Liang, Y.; Niu, Y. Rosmarinic acid attenuates beta-amyloid-induced oxidative stress via Akt/GSK-3beta/Fyn-mediated Nrf2 activation in PC12 cells. Free Radic. Biol. Med. 2018, 120, 114–123. [Google Scholar] [CrossRef]
- Winter, A.N.; Ross, E.K.; Khatter, S.; Miller, K.; Linseman, D.A. Chemical basis for the disparate neuroprotective effects of the anthocyanins, callistephin and kuromanin, against nitrosative stress. Free Radic. Biol. Med. 2017, 103, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Vauzour, D.; Rodriguez-Mateos, A.; Corona, G.; Oruna-Concha, M.J.; Spencer, J.P. Polyphenols and human health: Prevention of disease and mechanisms of action. Nutrients 2010, 2, 1106–1131. [Google Scholar] [CrossRef]
- Yang, E.J.; Mahmood, U.; Kim, H.; Choi, M.; Choi, Y.; Lee, J.P.; Cho, J.Y.; Hyun, J.W.; Kim, Y.S.; Chang, M.J.; et al. Phloroglucinol ameliorates cognitive impairments by reducing the amyloid beta peptide burden and pro-inflammatory cytokines in the hippocampus of 5XFAD mice. Free Radic. Biol. Med. 2018, 126, 221–234. [Google Scholar] [CrossRef] [PubMed]
- Williams, R.J.; Spencer, J.P. Flavonoids, cognition, and dementia: Actions, mechanisms, and potential therapeutic utility for Alzheimer disease. Free Radic. Biol. Med. 2012, 52, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, E.; Ahmad, A.; Singh, S.; Arshad, M.; Khan, A.H.; Khan, R.H. A mechanistic approach for islet amyloid polypeptide aggregation to develop anti-amyloidogenic agents for type-2 diabetes. Biochime 2011, 93, 793–805. [Google Scholar] [CrossRef] [PubMed]
- Francioso, A.; Punzi, P.; Boffi, A.; Lori, C.; Martire, S.; Giordano, C.; D’Erme, M.; Mosca, L. Beta-sheet interfering molecules acting against beta-amyloid aggregation and fibrillogenesis. Bioorgan. Med. Chem. 2015, 23, 1671–1683. [Google Scholar] [CrossRef]
- Pahnke, J.; Frohlich, C.; Paarmann, K.; Krohn, M.; Bogdanovic, N.; Arsland, D.; Winblad, B. Cerebral ABC transporter-common mechanisms may modulate neurodegenerative diseases and depression in elderly subjects. Arch. Med. Res. 2014, 45, 738–743. [Google Scholar] [CrossRef]
- Brenn, A.; Grube, M.; Jedlitschky, G.; Fischer, A.; Strohmeier, B.; Eiden, M.; Keller, M.; Groschup, M.H.; Vogelgesang, S. St. John’s Wort reduces beta-amyloid accumulation in a double transgenic Alzheimer’s disease mouse model-role of P-glycoprotein. Brain Pathol. 2014, 24, 18–24. [Google Scholar] [CrossRef]
- Hofrichter, J.; Krohn, M.; Schumacher, T.; Lange, C.; Feistel, B.; Walbroel, B.; Pahnke, J. Sideritis spp. Extracts Enhance Memory and Learning in Alzheimer’s beta-Amyloidosis Mouse Models and Aged C57Bl/6 Mice. J. Alzheimer’s Dis. JAD 2016, 53, 967–980. [Google Scholar] [CrossRef]
- Nicolia, V.; Cavallaro, R.A.; Lopez-Gonzalez, I.; Maccarrone, M.; Scarpa, S.; Ferrer, I.; Fuso, A. DNA Methylation Profiles of Selected Pro-Inflammatory Cytokines in Alzheimer Disease. J. Neuropathol. Exp. Neurol. 2017, 76, 27–31. [Google Scholar] [CrossRef]
- Sezgin, Z.; Dincer, Y. Alzheimer’s disease and epigenetic diet. Neurochem. Int. 2014, 78, 105–116. [Google Scholar] [CrossRef] [PubMed]
- Latta, C.H.; Brothers, H.M.; Wilcock, D.M. Neuroinflammation in Alzheimer’s Disease; a Source of Heterogeneity and Target for Personalized Therapy. Neuroscience 2015, 302, 103–111. [Google Scholar] [CrossRef]
- Rubio-Perez, J.M.; Morillas-Ruiz, J.M. A Review: Inflammatory Process in Alzheimer’s Disease, Role of Cytokines. Sci. World J. 2012. [Google Scholar] [CrossRef] [PubMed]
- Engelhart, M.J.; Geerlings, M.I.; Ruitenberg, A.; van Swieten, J.C.; Holman, A.; Witteman, J.C.M.; Breteler, M.M.B. Dietary intake of antioxidants and risk of Alzheimer disease. JAMA-J. Am. Med. Assoc. 2002, 287, 3223–3229. [Google Scholar] [CrossRef]
- Ringman, J.M.; Frautschy, S.A.; Teng, E.; Begum, A.N.; Bardens, J.; Beigi, M.; Gylys, K.H.; Badmaev, V.; Heath, D.D.; Apostolova, L.G.; et al. Oral curcumin for Alzheimer’s disease: Tolerability and efficacy in a 24-week randomized, double blind, placebo-controlled study. Alzheimers Res. Ther. 2012, 4, 43. [Google Scholar] [CrossRef] [PubMed]
- Mazzanti, G.; Di Giacomo, S. Curcumin and Resveratrol in the Management of Cognitive Disorders: What Is the Clinical Evidence? Molecules 2016, 21, 1243. [Google Scholar] [CrossRef]
- Omar, S.H.; Scott, C.J.; Hamlin, A.S.; Obied, H.K. The protective role of plant biophenols in mechanisms of Alzheimer’s disease. J. Nutr. Biochem. 2017, 47, 1–20. [Google Scholar] [CrossRef]
- Biancucci, M.; Mattioli, R.; Moubayidin, L.; Sabatini, S.; Costantino, P.; Trovato, M. Proline affects the size of the root meristematic zone in Arabidopsis. BMC Plant Biol. 2015, 15, 263. [Google Scholar] [CrossRef]
- Brunetti, P.; Zanella, L.; De Paolis, A.; Di Litta, D.; Cecchetti, V.; Falasca, G.; Barbieri, M.; Altamura, M.M.; Costantino, P.; Cardarelli, M. Cadmium-inducible expression of the ABC-type transporter AtABCC3 increases phytochelatin-mediated cadmium tolerance in Arabidopsis. J. Exp. Bot. 2015, 66, 3815–3829. [Google Scholar] [CrossRef] [Green Version]
- Von Roepenack-Lahaye, E.; Degenkolb, T.; Zerjeski, M.; Franz, M.; Roth, U.; Wessjohann, L.; Schmidt, J.; Scheel, D.; Clemens, S. Profiling of Arabidopsis secondary metabolites by capillary liquid chromatography coupled to electrospray ionization quadrupole time-of-flight mass spectrometry. Plant Physiol. 2004, 134, 548–559. [Google Scholar] [CrossRef]
- Beale, M.H.; Sussman, M.R. Metabolomics of Arabidopsis thaliana. In Annual Plant Reviews; Hall, R.D., Ed.; Wiley-Blackwell: Leiden, The Netherlands, 2011; Volume 43, pp. 157–180. [Google Scholar]
- Li, J.Y.; Oulee, T.M.; Raba, R.; Amundson, R.G.; Last, R.L. Arabidopsis Flavonoid Mutants Are Hypersensitive to Uv-B Irradiation. Plant Cell 1993, 5, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Ryan, K.G.; Swinny, E.E.; Winefield, C.; Markham, K.R. Flavonoids and UV photoprotection in Arabidopsis mutants. Z. Naturforsch. C 2001, 56, 745–754. [Google Scholar] [CrossRef] [PubMed]
- Onkokesung, N.; Reichelt, M.; van Doorn, A.; Schuurink, R.C.; van Loon, J.J.A.; Dicke, M. Modulation of flavonoid metabolites in Arabidopsis thaliana through overexpression of the MYB75 transcription factor: Role of kaempferol-3,7-dirhamnoside in resistance to the specialist insect herbivore Pieris brassicae. J. Exp. Bot. 2014, 65, 2203–2217. [Google Scholar] [CrossRef] [PubMed]
- Routaboul, J.M.; Kerhoas, L.; Debeaujon, I.; Pourcel, L.; Caboche, M.; Einhorn, J.; Lepiniec, L. Flavonoid diversity and biosynthesis in seed of Arabidopsis thaliana. Planta 2006, 224, 96–107. [Google Scholar] [CrossRef] [PubMed]
- Ghura, S.; Tai, L.; Zhao, M.; Collins, N.; Che, C.T.; Warpeha, K.M.; LaDu, M.J. Arabidopsis thaliana extracts optimized for polyphenols production as potential therapeutics for the APOE-modulated neuroinflammation characteristic of Alzheimer’s disease in vitro. Sci. Rep. 2016, 6, 29364. [Google Scholar] [CrossRef] [PubMed]
- Martire, S.; Fuso, A.; Mosca, L.; Forte, E.; Correani, V.; Fontana, M.; Scarpa, S.; Maras, B.; d’Erme, M. Bioenergetic Impairment in Animal and Cellular Models of Alzheimer’s Disease: PARP-1 Inhibition Rescues Metabolic Dysfunctions. J. Alzheimers Dis. 2016, 54, 307–324. [Google Scholar] [CrossRef] [PubMed]
- Raha, S.; Lee, H.J.; Yumnam, S.; Hong, G.E.; Saralamma, V.V.G.; Ha, Y.L.; Kim, J.O.; Kim, Y.S.; Heo, J.D.; Lee, S.J.; et al. Vitamin D-2 suppresses amyloid-beta 25–35 induced microglial activation in BV2 cells by blocking the NF-kappa B inflammatory signaling pathway. Life Sci. 2016, 161, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Awasaki, T.; Lai, S.L.; Ito, K.; Lee, T. Organization and Postembryonic Development of Glial Cells in the Adult Central Brain of Drosophila. J. Neurosci. 2008, 28, 13742–13753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akiyama, H.; Barger, S.; Barnum, S.; Bradt, B.; Bauer, J.; Cole, G.M.; Cooper, N.R.; Eikelenboom, P.; Emmerling, M.; Fiebich, B.L.; et al. Inflammation and Alzheimer’s disease. Neurobiol. Aging 2000, 21, 383–421. [Google Scholar] [CrossRef]
- Masci, A.; Mattioli, R.; Costantino, P.; Baima, S.; Morelli, G.; Punzi, P.; Giordano, C.; Pinto, A.; Donini, L.M.; d’Erme, M.; et al. Neuroprotective Effect of Brassica oleracea Sprouts Crude Juice in a Cellular Model of Alzheimer’s Disease. Oxid. Med. Cell. Longev. 2015. [Google Scholar] [CrossRef]
- Alonso-Blanco, C.; Aarts, M.G.M.; Bentsink, L.; Keurentjes, J.J.B.; Reymond, M.; Vreugdenhil, D.; Koornneef, M. What Has Natural Variation Taught Us about Plant Development, Physiology, and Adaptation? Plant Cell 2009, 21, 1877–1896. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weigel, D. Natural Variation in Arabidopsis: From Molecular Genetics to Ecological Genomics. Plant Physiol. 2012, 158, 2–22. [Google Scholar] [CrossRef] [PubMed]
- Routaboul, J.M.; Dubos, C.; Beck, G.; Marquis, C.; Bidzinski, P.; Loudet, O.; Lepiniec, L. Metabolite profiling and quantitative genetics of natural variation for flavonoids in Arabidopsis. J. Exp. Bot. 2012, 63, 3749–3764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.-Y.; Tan, M.-S.; Yu, J.-T.; Tan, L. Role of pro-inflammatory cytokines released from microglia in Alzheimer’s disease. Ann. Transl. Med. 2015, 3, 136. [Google Scholar]
- Cherry, J.D.; Olschowka, J.A.; O’Banion, M.K. Neuroinflammation and M2 microglia: The good, the bad, and the inflamed. J. Neuroinflamm. 2014, 11, 98. [Google Scholar] [CrossRef] [PubMed]
- Haack, M.; Lowinger, M.; Lippmann, D.; Kipp, A.; Pagnotta, E.; Iori, R.; Monien, B.H.; Glatt, H.; Brauer, M.N.; Wessjohann, L.A.; et al. Breakdown products of neoglucobrassicin inhibit activation of Nrf2 target genes mediated by myrosinase-derived glucoraphanin hydrolysis products. Biol. Chem. 2010, 391, 1281–1293. [Google Scholar] [CrossRef]
- Kobayashi, E.H.; Suzuki, T.; Funayama, R.; Nagashima, T.; Hayashi, M.; Sekine, H.; Tanaka, N.; Moriguchi, T.; Motohashi, H.; Nakayama, K.; et al. Nrf2 suppresses macrophage inflammatory response by blocking proinflammatory cytokine transcription. Nat. Commun. 2016, 7, 11624. [Google Scholar] [CrossRef] [Green Version]
- Wruck, C.J.; Streetz, K.; Pavic, G.; Gotz, M.E.; Tohidnezhad, M.; Brandenburg, L.-O.; Varoga, D.; Eickelberg, O.; Herdegen, T.; Trautwein, C.; et al. Nrf2 induces interleukin-6 (IL-6) expression via an antioxidant response element within the IL-6 promoter. J. Biol. Chem. 2011, 286, 4493–4499. [Google Scholar] [CrossRef]
- Finelli, A.; Kelkar, A.; Song, H.J.; Yang, H.; Konsolaki, M. A model for studying Alzheimer’s Abeta42-induced toxicity in Drosophila melanogaster. Mol. Cell. Neurosci. 2004, 26, 365–375. [Google Scholar] [CrossRef]
- Crowther, D.C.; Kinghorn, K.J.; Miranda, E.; Page, R.; Curry, J.A.; Duthie, F.A.; Gubb, D.C.; Lomas, D.A. Intraneuronal Abeta, non-amyloid aggregates and neurodegeneration in a Drosophila model of Alzheimer’s disease. Neuroscience 2005, 132, 123–135. [Google Scholar] [CrossRef]
- Iijima, K.; Iijima-Ando, K. Drosophila models of Alzheimer’s amyloidosis: The challenge of dissecting the complex mechanisms of toxicity of amyloid-beta 42. J. Alzheimers Dis. 2008, 15, 523–540. [Google Scholar] [CrossRef] [PubMed]
- Prussing, K.; Voigt, A.; Schulz, J.B. Drosophila melanogaster as a model organism for Alzheimer’s disease. Mol. Neurodegener. 2013, 8, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ping, Y.; Hahm, E.T.; Waro, G.; Song, Q.; Vo-Ba, D.A.; Licursi, A.; Bao, H.; Ganoe, L.; Finch, K.; Tsunoda, S. Linking A beta 42-Induced Hyperexcitability to Neurodegeneration, Learning and Motor Deficits, and a Shorter Lifespan in an Alzheimer’s Model. PLoS Genet. 2015, 11, e1005025. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Del-Rio, M.; Velez-Pardo, C. Alzheimer’s Disease, Drosophila melanogaster and Polyphenols. Adv. Exp. Med. Biol. 2015, 863, 21–53. [Google Scholar] [PubMed]
- Kong, Y.; Li, K.; Fu, T.; Wan, C.; Zhang, D.; Song, H.; Zhang, Y.; Liu, N.; Gan, Z.; Yuan, L. Quercetin ameliorates Abeta toxicity in Drosophila AD model by modulating cell cycle-related protein expression. Oncotarget 2016, 7, 67716–67731. [Google Scholar] [CrossRef]
- Caesar, I.; Jonson, M.; Nilsson, K.P.; Thor, S.; Hammarstrom, P. Curcumin promotes A-beta fibrillation and reduces neurotoxicity in transgenic Drosophila. PLoS ONE 2012, 7, e31424. [Google Scholar] [CrossRef] [PubMed]
- Beg, T.; Jyoti, S.; Naz, F.; Rahul; Ali, F.; Ali, S.K.; Reyad, A.M.; Siddique, Y.H. Protective Effect of Kaempferol on the Transgenic Drosophila Model of Alzheimer’s Disease. CNS Neurol. Disord. Drug Targets 2018, 17, 421–429. [Google Scholar] [CrossRef]
- Atherton, E.; Sheppard, R.C. Solid Phase Synthesis—A Practical Approach; IRL Press at Oxford University Press: Cary, NC, USA, 1989; p. 203. [Google Scholar]
- Singleton, V.L.; Rossi, J.A., Jr. Colorimetry of Total Phenolics with Phosphomolybdic-Phosphotungstic Acid Reagents. Am. J. Enol. Vitic. 1965, 16, 144–158. [Google Scholar]
- Giusti, M.M.; Wrolstad, R.E. Characterization and measurement of anthocyanins by UV-visible spectroscopy. In Current Protocols in Food Analytical Chemistry; Wrolstad, R.E., Schwartz, S.J., Eds.; Wiley: New York, NY, USA, 2001; pp. F1.2.1–F1.2.13. [Google Scholar]
- Gaikwad, A.; Long, D.J.; Stringer, J.L.; Jaiswal, A.K. In vivo role of NAD(P)H:quinone oxidoreductase 1 (NQO1) in the regulation of intracellular redox state and accumulation of abdominal adipose tissue. J. Biol. Chem. 2001, 276, 22559–22564. [Google Scholar] [CrossRef]
- Feiguin, F.; Godena, V.K.; Romano, G.; D’Ambrogio, A.; Klima, R.; Baralle, F.E. Depletion of TDP-43 affects Drosophila motoneurons terminal synapsis and locomotive behavior. FEBS Lett. 2009, 583, 1586–1592. [Google Scholar] [CrossRef]





| Peak | Compound | Molecular Mass (Calculated) | Molecular Formula | λ Max (nm) | MS1 [M-H]− m/z | MS1 [M+H]+ m/z | MS2 [M-H]− m/z | MS2 [M+H]+ m/z |
|---|---|---|---|---|---|---|---|---|
| 1 * | Caffeic acid | 180.06 | C9H8O4 | 326 | 179 | |||
| 2 | Quercetin-rhamnoside-hexoside | 610.16 | C27H30O16 | 350 | 609 | 611 | 447, 301 | 449, 303 |
| 3 | Kaempferol-3-O-glucoside-7-O-rhamnoside | 594.16 | C27H30O15 | 351 | 593 | 595 | 431, 285 | 433, 287 |
| 4 | Quercetin-di-rhamnoside | 594.16 | C27H30O15 | 350 | 593 | 595 | 447, 301 | 449, 303 |
| 5 | Isorhamnetin-hexoside-rhamnoside | 624.17 | C28H32O16 | 345 | 623 | 625 | 315 | 463, 317 |
| 6 | Kaempferol-3, 7-di-O-rhamnoside | 578.17 | C27H30O14 | 351 | 577 | 579 | 433, 287 | |
| 7 * | Synapic acid | 224.06 | C11H12O5 | 327 | 223 | |||
| 8–9 | di-O-sinapoyl-β-glucose (isomers) | 592.17 | C28H32O14 | 327 | 591 | 367, 349, 223 | ||
| 10 * | Luteolin | 286.05 | C15H10O6 | 350 | 285 | 287 |
| Gene | Primer Sequence | |
|---|---|---|
| FORWARD | REVERSE | |
| Heme oxygenase-1 (HO-1) | AGGAGGTACACATCCAAGCC | TACAAGGAAGCCATCACCAG |
| Interleukin 6 (IL-6) | AAGCTGGAGTCACAGAAGGAG | GGTTTGCCGAGTAGATCTCAA |
| Interleukin 1-β (IL-1β) | TTCGTGAATGAGCAGACAGC | CCATGGTTTCTTGTGACCCT |
| TNF-α | TGGCCTCTCTACCTTGTTGC | GGGAGCAGAGGTTCAGTGAT |
| Interleukin 4 (IL-4) | TGTACCAGGAGCCATATCCA | TTCTTCGTTGCTGTGAGGAC |
| Interleukin 10 (IL-10) | CCCAGAAATCAAGGAGCATT | TCACTCTTCACCTGCTCCAC |
| Interleukin 13 (IL-13) | AGCATGGTATGGAGTGTGGA | TTGCAATTGGAGATGTTGGT |
| Ribosomal protein S27a (RPS27A) | AGAGGCTGATCTTTGCTGGT | ACCAGATGAAGGGTGGACTC |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mattioli, R.; Francioso, A.; d’Erme, M.; Trovato, M.; Mancini, P.; Piacentini, L.; Casale, A.M.; Wessjohann, L.; Gazzino, R.; Costantino, P.; et al. Anti-Inflammatory Activity of A Polyphenolic Extract from Arabidopsis thaliana in In Vitro and In Vivo Models of Alzheimer’s Disease. Int. J. Mol. Sci. 2019, 20, 708. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20030708
Mattioli R, Francioso A, d’Erme M, Trovato M, Mancini P, Piacentini L, Casale AM, Wessjohann L, Gazzino R, Costantino P, et al. Anti-Inflammatory Activity of A Polyphenolic Extract from Arabidopsis thaliana in In Vitro and In Vivo Models of Alzheimer’s Disease. International Journal of Molecular Sciences. 2019; 20(3):708. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20030708
Chicago/Turabian StyleMattioli, Roberto, Antonio Francioso, Maria d’Erme, Maurizio Trovato, Patrizia Mancini, Lucia Piacentini, Assunta Maria Casale, Ludger Wessjohann, Roberta Gazzino, Paolo Costantino, and et al. 2019. "Anti-Inflammatory Activity of A Polyphenolic Extract from Arabidopsis thaliana in In Vitro and In Vivo Models of Alzheimer’s Disease" International Journal of Molecular Sciences 20, no. 3: 708. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20030708