Exosome-Mediated Delivery of Inducible miR-423-5p Enhances Resistance of MRC-5 Cells to Rabies Virus Infection
Abstract
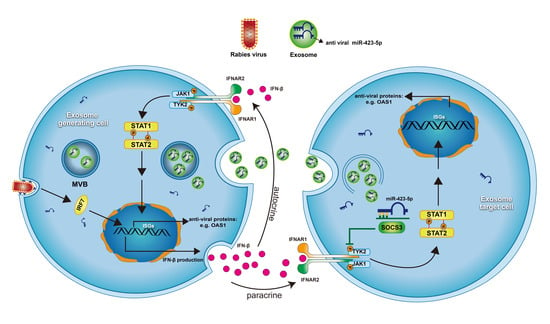
:1. Introduction
2. Results
2.1. Blocking Exosomal Release Promotes RABV Infection in MRC-5 Cells
2.2. RABV Infection Alters miRNA Expression Patterns in Exosomes
2.3. miR-423-5p Targets Suppressor of Cytokine Signaling 3 (SOCS3)
2.4. miR-423-5p Feedback Inhibits RABV Replication in MRC-5 Cells
2.5. RABV Infection Induces Up-Regulation of IFN-β in MRC-5 Cells
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Exosome Isolation
4.3. Exosome Labeling and Uptake Analysis
4.4. Small RNA Deep Sequencing Analysis
4.5. Transient Transfection of siRNAs and miRNAs
4.6. Luciferase Reporter Assays
4.7. RNA Extraction and Quantitative RT-PCR
4.8. Western Blotting
4.9. Transmission Electron Microscopy
4.10. Nanoparticle Tracking Analysis
4.11. Immunofluorescence Microscopy
4.12. Statistical Analysis
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
Abbreviations
| DAPI | 4′,6-diamidino-2-phenylindole |
| DiO | dioctadecyloxacarbocyanine perchlorate |
| Exo | exosome |
| HCV | hepatitis C virus |
| HDCs | human diploid cells |
| IFN | interferon |
| JAK-SATA | Janus Kinase-Signal Transducers and Activators of Transcription |
| KSHV | Kaposi’s sarcoma-associated herpesvirus |
| miRNA | microRNA |
| MRC-5 cells | human embryonic lung fibroblast cells |
| Mut | mutant |
| MVB | multivesicular body |
| NTA | nanoparticle tracking analysis |
| RABV | rabies virus |
| siRNA | small interfering RNA |
| SOCS3 | suppressor of cytokine signaling 3 |
| SSI | STAT-induced STAT inhibitor |
| TEM | transmission electron microscopy |
| WT | wild-type |
References
- Tordo, N.; Poch, O.; Ermine, A.; Keith, G.; Rougeon, F. Walking along the rabies genome: Is the large G-L intergenic region a remnant gene? Proc. Natl. Acad. Sci. USA 1986, 83, 3914–3918. [Google Scholar] [CrossRef] [PubMed]
- GBD 2015 Mortality and Causes of Death Collaborators. Global, regional, and national life expectancy, all-cause mortality, and cause-specifi c mortality for 249 causes of death, 1980–2015: A systematic analysis for the Global Burden of Disease Study 2015. Lancet 2016, 388, 1459–1544. [Google Scholar] [CrossRef]
- Hicks, D.J.; Fooks, A.R.; Johnson, N. Developments in rabies vaccines. Clin. Exp. Immunol. 2012, 169, 199–204. [Google Scholar] [CrossRef] [Green Version]
- Kondo, A. Growth characteristics of rabies virus in primary chicken embryo cells. Virology 1965, 27, 199–204. [Google Scholar] [CrossRef]
- Lavender, J.F.; Van Frank, R.M. Zonal-centrifuged purifed duck embryo cell culture rabies vaccine for human vaccination. Appl. Microbiol. 1971, 22, 358–365. [Google Scholar]
- Majer, M.; Hilfenhaus, A.J.; Mauler, R.; Lehmann, H.G.; Hennessen, W.; Kuwert, E.K. A comparison of the Pasteur and Pitman-Moore strains of rabies virus for the production of rabies vaccine in human diploid cells. J. Biol. Standard. 1977, 5, 249–256. [Google Scholar] [CrossRef]
- Lin, F.; Lu, L. Rabies vaccine production in animal cell cultures. J. Infect. Dis. 1983, 147, 467–473. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Smith, T.G.; Rupprecht, C.E. From brain passage to cell adaptation: The road of human rabies vaccine development. Expert Rev. Vaccines 2011, 10, 1597–1608. [Google Scholar] [CrossRef]
- Jacobs, J.P. Characteristics of a human diploid cell designated MRC-5. Nature 1970, 227, 168–170. [Google Scholar] [CrossRef] [PubMed]
- Maria, I.T.; Melitta, S.; Ursula, H.; Harold, C.; Monique, L. The Neural Cell Adhesion Molecule Is a Receptor for Rabies Virus. J. Virol. 1998, 7181–7190. [Google Scholar]
- Beutler, B.; Jiang, Z.; Georgel, P.; Crozat, K.; Croker, B.; Rutschmann, S.; Du, X.; Hoebe, K. Genetic analysis of host resistance: Toll-like receptor signaling and immunity at large. Annu. Rev. Immunol. 2006, 24, 353–389. [Google Scholar] [CrossRef]
- Kim, B.H.; Shenoy, A.R.; Kumar, P.; Bradfield, C.J.; MacMicking, J.D. IFN-inducible GTPases in host cell defense. Cell Host Microbe 2012, 12, 432–444. [Google Scholar] [CrossRef] [PubMed]
- Assil, S.; Webster, B.; Dreux, M. Regulation of the Host Antiviral State by Intercellular Communications. Viruses 2015, 7, 4707–4733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Randall, R.E.; Goodbourn, S. Interferons and viruses: An interplay between induction, signalling, antiviral responses and virus countermeasures. J. General Virol. 2008, 89, 1–47. [Google Scholar] [CrossRef]
- Antwi-Baffour, S.S. Molecular characterisation of plasma membrane-derived vesicles. J. Biomed. Sci. 2015, 22, 68. [Google Scholar] [CrossRef]
- Tkach, M.; Thery, C. Communication by Extracellular Vesicles: Where We Are and Where We Need to Go. Cell 2016, 164, 1226–1232. [Google Scholar] [CrossRef] [PubMed]
- Kalra, H.; Drummen, G.P.; Mathivanan, S. Focus on Extracellular Vesicles: Introducing the Next Small Big Thing. Int. J. Mol. Sci. 2016, 17, 170. [Google Scholar] [CrossRef] [PubMed]
- Schorey, J.S.; Cheng, Y.; Singh, P.P.; Smith, V.L. Exosomes and other extracellular vesicles in host-pathogen interactions. EMBO Rep. 2015, 16, 24–43. [Google Scholar] [CrossRef] [PubMed]
- Raab-Traub, N.; Dittmer, D.P. Viral effects on the content and function of extracellular vesicles. Nat. Rev. Microbiol. 2017, 15, 559–572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petrik, J. Immunomodulatory effects of exosomes produced by virus-infected cells. Transf. Apher. Sci. 2016, 55, 84–91. [Google Scholar] [CrossRef]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [Green Version]
- Ramakrishnaiah, V.; Thumann, C.; Fofana, I.; Habersetzer, F.; Pan, Q.; de Ruiter, P.E.; Willemsen, R.; Demmers, J.A.; Stalin Raj, V.; Jenster, G.; et al. Exosome-mediated transmission of hepatitis C virus between human hepatoma Huh7.5 cells. Proc. Natl. Acad. Sci. USA 2013, 110, 13109–13113. [Google Scholar] [CrossRef] [Green Version]
- Lenassi, M.; Cagney, G.; Liao, M.; Vaupotic, T.; Bartholomeeusen, K.; Cheng, Y.; Krogan, N.J.; Plemenitas, A.; Peterlin, B.M. HIV Nef is secreted in exosomes and triggers apoptosis in bystander CD4+ T cells. Traffic 2010, 11, 110–122. [Google Scholar] [CrossRef] [PubMed]
- Pleet, M.L.; DeMarino, C.; Lepene, B.; Aman, M.J.; Kashanchi, F. The Role of Exosomal VP40 in Ebola Virus Disease. DNA Cell Biol. 2017, 36, 243–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, X.; Odenthal, M.; Fries, J.W. Exosomes as miRNA Carriers: Formation-Function-Future. Int. J. Mol. Sci. 2016, 17, 2028. [Google Scholar] [CrossRef] [PubMed]
- Pegtel, D.M.; Cosmopoulos, K.; Thorley-Lawson, D.A.; van Eijndhoven, M.A.; Hopmans, E.S.; Lindenberg, J.L.; de Gruijl, T.D.; Wurdinger, T.; Middeldorp, J.M. Functional delivery of viral miRNAs via exosomes. Proc. Natl. Acad. Sci. USA 2010, 107, 6328–6333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vojtech, L.; Woo, S.; Hughes, S.; Levy, C.; Ballweber, L.; Sauteraud, R.P.; Strobl, J.; Westerberg, K.; Gottardo, R.; Tewari, M.; et al. Exosomes in human semen carry a distinctive repertoire of small non-coding RNAs with potential regulatory functions. Nucleic Acids Res. 2014, 42, 7290–7304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalamvoki, M.; Du, T.; Roizman, B. Cells infected with herpes simplex virus 1 export to uninfected cells exosomes containing STING, viral mRNAs, and microRNAs. Proc. Natl. Acad. Sci. USA 2014, 111, E4991–E4996. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Liu, K.; Liu, Y.; Xu, Y.; Zhang, F.; Yang, H.; Liu, J.; Pan, T.; Chen, J.; Wu, M.; et al. Exosomes mediate the cell-to-cell transmission of IFN-alpha-induced antiviral activity. Nat. Immunol. 2013, 14, 793–803. [Google Scholar] [CrossRef] [PubMed]
- Giugliano, S.; Kriss, M.; Golden-Mason, L.; Dobrinskikh, E.; Stone, A.E.L.; Soto-Gutierrez, A.; Mitchell, A.; Khetani, S.R.; Yamane, D.; Stoddard, M.; et al. Hepatitis C Virus Infection Induces Autocrine Interferon Signaling by Human Liver Endothelial Cells and Release of Exosomes, Which Inhibits Viral Replication. Gastroenterology 2015, 148, 392–402.e13. [Google Scholar] [CrossRef]
- Narayanan, A.; Jaworski, E.; Van Duyne, R.; Iordanskiy, S.; Guendel, I.; Das, R.; Currer, R.; Sampey, G.; Chung, M.; Kehn-Hall, K.; et al. Exosomes derived from HTLV-1 infected cells contain the viral protein Tax. Retrovirology 2014, 11, O46. [Google Scholar] [CrossRef]
- Ariza, M.E.; Rivailler, P.; Glaser, R.; Chen, M.; Williams, M.V. Epstein-Barr virus encoded dUTPase containing exosomes modulate innate and adaptive immune responses in human dendritic cells and peripheral blood mononuclear cells. PLoS ONE 2013, 8, e69827. [Google Scholar] [CrossRef] [PubMed]
- Honegger, A.; Schilling, D.; Bastian, S.; Sponagel, J.; Kuryshev, V.; Sultmann, H.; Scheffner, M.; Hoppe-Seyler, K.; Hoppe-Seyler, F. Dependence of intracellular and exosomal microRNAs on viral E6/E7 oncogene expression in HPV-positive tumor cells. PLoS Pathog. 2015, 11, e1004712. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Zhou, Q.; Xie, Y.; Zuo, L.; Zhu, F.; Lu, J. Extracellular vesicles: Novel vehicles in herpesvirus infection. Virol. Sin. 2017, 32, 349–356. [Google Scholar] [CrossRef]
- Zhou, Y.; Wang, X.; Sun, L.; Zhou, L.; Ma, T.C.; Song, L.; Wu, J.G.; Li, J.L.; Ho, W.Z. Toll-like receptor 3-activated macrophages confer anti-HCV activity to hepatocytes through exosomes. FASEB J. 2016, 30, 4132–4140. [Google Scholar] [CrossRef]
- Kouwaki, T.; Okamoto, M.; Tsukamoto, H.; Fukushima, Y.; Oshiumi, H. Extracellular Vesicles Deliver Host and Virus RNA and Regulate Innate Immune Response. Int. J. Mol. Sci. 2017, 18, 666. [Google Scholar] [CrossRef]
- Bellingham, S.A.; Coleman, B.M.; Hill, A.F. Small RNA deep sequencing reveals a distinct miRNA signature released in exosomes from prion-infected neuronal cells. Nucleic Acids Res. 2012, 40, 10937–10949. [Google Scholar] [CrossRef] [Green Version]
- Verweij, F.J.; van Eijndhoven, M.A.; Middeldorp, J.; Pegtel, D.M. Analysis of viral microRNA exchange via exosomes in vitro and in vivo. Methods Mol. Biol. 2013, 1024, 53–68. [Google Scholar] [PubMed]
- Yogev, O.; Henderson, S.; Hayes, M.J.; Marelli, S.S.; Ofir-Birin, Y.; Regev-Rudzki, N.; Herrero, J.; Enver, T. Herpesviruses shape tumour microenvironment through exosomal transfer of viral microRNAs. PLoS Pathog. 2017, 13, e1006524. [Google Scholar] [CrossRef] [PubMed]
- Chugh, P.E.; Sin, S.H.; Ozgur, S.; Henry, D.H.; Menezes, P.; Griffith, J.; Eron, J.J.; Damania, B.; Dittmer, D.P. Systemically circulating viral and tumor-derived microRNAs in KSHV-associated malignancies. PLoS Pathog. 2013, 9, e1003484. [Google Scholar] [CrossRef]
- Song, M.M.; Shuai, K. The suppressor of cytokine signaling (SOCS) 1 and SOCS3 but not SOCS2 proteins inhibit interferon-mediated antiviral and antiproliferative activities. J. Biol. Chem. 1998, 273, 35056–35062. [Google Scholar] [CrossRef] [PubMed]
- Kreuzer, R.S.; Bekurtz, J.C.; Arends, D.; Bortfeldt, R.; Kutz, L.B.; Sharbati, S.; Einspanier, R.; Brockmann, G.A. Feeding of Enterococcus faecium NCIMB 10415 Leads to Intestinal miRNA-423-5p-Induced Regulation of Immune-Relevant Genes. Appl. Environ. Microbiol. 2016, 82, 2263–2269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.G.; Wang, F. MiR-663a/MiR-423-5p are involved in the pathogenesis of lupus nephritis via modulating the activation of NF-κB by targeting TNIP2. Am. J. Transl. Res. 2017, 9, 3796–3803. [Google Scholar] [PubMed]
- Théry, C.; Amigorena, S.; Raposo, G.; Clayton, A. Isolation and Characterization of Exosomes from Cell Culture Supernatants and Biological Fluids; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2006; p. Unit 3.22. [Google Scholar]
- Liang, G.; Kan, S.; Zhu, Y.; Feng, S.; Feng, W.; Gao, S. Engineered exosome-mediated delivery of functionally active miR-26a and its enhanced suppression effect in HepG2 cells. Int. J. Nanomed. 2018, 13, 585–599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Liu, Y.; Zhang, S.; Wang, Y.; Zhao, J.; Miao, F.; Hu, R. A SYBR-green I quantitative real-time reverse transcription-PCR assay for rabies viruses with different virulence. Virol. Sin. 2014, 29, 131–132. [Google Scholar] [CrossRef] [PubMed]
- Remoli, M.E.; Giacomini, E.; Lutfalla, G.; Dondi, E.; Orefici, G.; Battistini, A.; Uze, G.; Pellegrini, S.; Coccia, E.M. Selective Expression of Type I IFN Genes in Human Dendritic Cells Infected with Mycobacterium tuberculosis. J. Immunol. 2002, 169, 366–374. [Google Scholar] [CrossRef] [PubMed]





| Gene | Forward Primer (5′-3′) | Reverse Primer (5′-3′) |
|---|---|---|
| Rabies virus N protein | CAAGATGTGTGCYAAYTGGAG | AGCCCTGGTTCGAACATTCT |
| IFN-β | GTCTCCTCCAAATTGCTCTC | ACAGGAGCTTCTGACACTGA |
| STAT1 | TTCTGTGTCTGAAGTGTAAGTGAA | TAACACGGGGATCTCAACAAGTTC |
| OAS1 | AGAAGGCAGCTCACGAAACC | CCACCACCCAAGTTTCCTGTA |
| IRF7 | GAGCCCTTACCTCCCCTGTTAT | CCACTGCAGCCCCTCATAG |
| 18S | CTTAGAGGGACAAGTGGCG | ACGCTGAGCCAGTCAGTGTA |
| miR-423-5p | GGGGTGAGGGGCAGAGAG | universal reverse primers |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Teng, Y.; Zhao, G.; Li, F.; Hou, A.; Sun, B.; Kong, W.; Gao, F.; Cai, L.; Jiang, C. Exosome-Mediated Delivery of Inducible miR-423-5p Enhances Resistance of MRC-5 Cells to Rabies Virus Infection. Int. J. Mol. Sci. 2019, 20, 1537. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20071537
Wang J, Teng Y, Zhao G, Li F, Hou A, Sun B, Kong W, Gao F, Cai L, Jiang C. Exosome-Mediated Delivery of Inducible miR-423-5p Enhances Resistance of MRC-5 Cells to Rabies Virus Infection. International Journal of Molecular Sciences. 2019; 20(7):1537. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20071537
Chicago/Turabian StyleWang, Jingyu, Yawei Teng, Guanshu Zhao, Fang Li, Ali Hou, Bo Sun, Wei Kong, Feng Gao, Linjun Cai, and Chunlai Jiang. 2019. "Exosome-Mediated Delivery of Inducible miR-423-5p Enhances Resistance of MRC-5 Cells to Rabies Virus Infection" International Journal of Molecular Sciences 20, no. 7: 1537. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20071537