Recombinant Adeno-Associated Viral Vectors (rAAV)-Vector Elements in Ocular Gene Therapy Clinical Trials and Transgene Expression and Bioactivity Assays
Abstract
:1. Introduction
1.1. Why Viral Vector-Based Gene Augmentation Therapy for Ocular Diseases?
1.2. rAAV Gene Therapy for Ocular Diseases—Advantages and Disadvantages
2. Ocular rAAV Vector-Based Therapies in Clinical Trials
3. Discovery of Cell-Specific Promoters for Ocular Gene Therapy
3.1. Core Promoters in rAAV-Vectors
3.2. Ubiquitous Promoters in rAAV-Vectors
3.3. Bicistronic and Tricistronic Promoters in rAAV-Vectors
3.4. Retina-Specific Promoters
3.5. Small Nuclear RNA (snRNA) Promoters
3.6. WPRE, Introns, miRNAs, and Other Elements in a rAAV-Gene Cassette
3.7. Polyadenylation Sequences in rAAV-Gene Cassette
3.8. rAAV Vector Cassettes and Inducible Promoters
4. Optimizing Genes for rAAV Vector Therapies (Minigenes, Dual/Triple rAAV-Vector, ITRs)
4.1. Intron Removal, Exon Removal, Surrogates, and Pathway-Modifying Therapies
4.2. Lentiviral and Dual/Triple rAAV Vectors
4.3. rAAV-Vectors Expressing CRISPR/Cas
4.4. Production and rAAV Vector Integration
4.4.1. Production: The Backbones and Bacterial Resistance Genes
4.4.2. Production: ITR Stabilization
4.4.3. rAAV Vector Integration into the Host Genome and Chromatin Association
4.5. Codon Optimization and Self-Complementary rAAVs
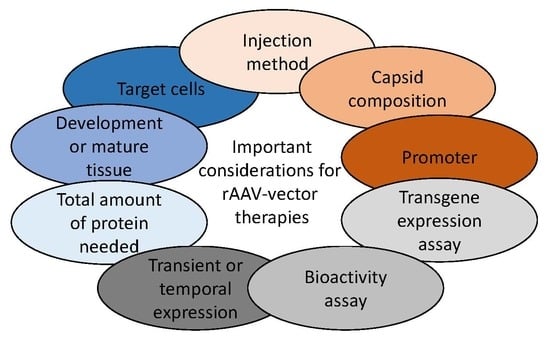
5. Transgene and Bioactivity Assays in Ocular Tissue
5.1. In Vitro Immortalized Epithelial Cell Lines for Transgene and Bioactivity Assays
5.2. In Vitro Immortalized Ocular Cell Lines for Transgene and Bioactivity Assays
5.3. In Vitro Differentiation of Human Induced Pluripotent Stem Cells (hiPSCs) to Retinal Pigment Epithelium (RPE) Cells
5.4. In Vitro Differentiation of Human Induced Pluripotent Stem Cells (hiPSCs) to Retinal Organoids for Transgene and Bioactivity Assays
5.5. Human Ex Vivo Retinal Culture for Transgene and Bioactivity Assays
5.6. In Vivo Studies for Transgene and Bioactivity Assays
5.6.1. Developmental Stage and rAAV Infection
5.6.2. rAAVs Overcoming Membranes in the Retina and the Retinal Disease State
5.6.3. Nonhuman Primate Studies and rAAV Infection
5.6.4. Cis-Regulatory Toxicity of rAAV Vectors In Vivo?
6. Concluding Remarks and Future Prospects
- The use of tyrosine-mutated rAAV2 capsids (AAV2-tYF; AAV2-7m8) increases retinal penetration and infection potentially replacing wild-type capsids (Section 2 and Section 5.6).
- The strong viral promoter CAG expresses the transgenes in the RPE for many years without being silenced [22] (Section 2, Section 3 and Section 5.6).
- Native promoters are more prone to differ in transgene expression levels in healthy-vs-disease states (Section 3.4 and Section 5).
- Promoters in general can greatly differ within in vivo/in vitro/ex vivo models, as well as across species (Section 3.4 and Section 5.6).
- Inducible promoters (riboswitches and dead-Cas9) offer exciting opportunities to control protein expression (Section 3.8).
- Surrogate gene (homolog/ortholog or synthetic) and minigene supplementation may circumvent cellular immunogenicity (Section 4.1).
- The rAAV production cell line and production cell line related impurities can influence the transduction efficiency in target tissue [44] (Section 2 and Section 4.4).
- Inverted terminal repeats of rAAVs are essential for high production yields but is not a prerequisite for the efficient transgene expression (Section 4.4.2).
- Genome integrations of rAAV vectors and the potential cell-toxic effect of genome integrations have been insufficiently studied in retinal tissue (Section 4.4.3).
- The rAAV infection pathway can vastly differ depending on the selected medium composition, the culturing technique/protocol, and the studied developmental state of the tissue (Section 5.6.1).
- The disease state strongly influences rAAV-vector penetration, potency, and tropism of the retina (Section 5.6.2).
7. Material and Methods
7.1. A Meta-Analysis on Pro-Viral Plasmids and Production Platforms for Ocular rAAV Therapies in Clinical Trials
7.2. Statistical Analysis
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
Abbreviations
| AAV | adeno-associated virus |
| AAV2-tYF | rAAV2-Y444F+Y500F+Y730F |
| Ad | adenovirus |
| AMD | age-related macular degeneration |
| AON | antisense oligonucleotides |
| BAA | biological activity assay |
| bGH | bovine growth hormone |
| CAG/CBA/pCAGGS | CMV early enhancer element, promoter of the first exon and the first intron of chicken β-actin gene, splice acceptor of the rabbit β-globin intron |
| CB7 | shortened CMV early enhancer element and a chicken β-actin promoter |
| CBh | CMV early enhancer element, a chicken β-actin promoter and a chimeric chicken β-Actin/minute virus of mice (MVM) viral protein (VP) intron |
| CB-SB | shortened CBA promoter containing a CMV early enhancer element |
| CMV | cytomegalovirus early enhancer element and promoter |
| EBV | Epstein Barr Virus |
| ERG | Electroretinogram (retinal function) |
| GCL | Ganglion cell layer |
| GMP | good manufacturing practice |
| hCAR | human cone arrestin |
| hCNGA3 | Human (cone PRC) Cyclic Nucleotide Gated Channel Subunit Alpha 3 |
| hCHM | Human CHM gene (encodes for the Rab escort protein 1 [REP1]) |
| hGH | human growth hormone |
| hGRK1/hRK | human G protein-coupled rhodopsin kinase 1 promoter |
| hiPSC | human induced pluripotent stem cells |
| hRPE65p | human retinal pigment epithelium-specific 65 kDa protein (RPE65) promoter |
| hVMD2 | human vitelliform macular dystrophy type 2 promoter (aka BEST1) |
| ILM | Inner Limiting Membrane |
| INL | Inner Nuclear Layer |
| IPL | Inner Plexiform Layer |
| IONs | inherited optic neuropathies |
| IRBP | interphotoreceptor retinoid-binding protein |
| IRDs | inherited retinal dystrophies |
| IRES | Internal ribosome entry site |
| ITR | palindromic inverted terminal repeats |
| ITR2 | palindromic inverted terminal repeats of AAV serotype 2 |
| LCA | Leber congenital amaurosis |
| LV | lentivirus |
| mCAR | mouse cone arrestin |
| mRNA | Messenger RNA |
| miRNA | microRNA |
| MGC | Müller glial cell |
| MVM | minute virus of mice viral protein (VP intron) |
| NMT | novel medical therapy |
| OKT | Optokinetic headtracking response |
| OLM | Outer Limiting Membrane |
| ONL | Outer Nuclear Layer |
| OPL | Outer Plexiform Layer |
| pA | polyadenylation sequence |
| PBGD | Porphobilinogen deaminase |
| PRC | photoreceptor |
| PRE | post-transcriptional regulatory element |
| rAAV | recombinant adeno-associated virus |
| rBG | rabbit β-globin |
| rHSV | replication-defective recombinant herpes virus |
| RPE | Retinal Pigment Epithelium |
| SpA | synthetic polyadenylation signal |
| sBHK | suspension-adapted Baby Hamster Kidney fibroblasts |
| scAAV | self-complementary recombinant adeno-associated virus |
| Sf9 | a clonal isolate of Spodoptera frugiperda (Fall Army worm) Sf21 cells |
| smCBA | truncated chimeric CMV/CBA promoter |
| ssAAV | single-stranded recombinant adeno-associated virus |
| SV40 | late simian virus 40 |
| TEA | transgene expression assay |
| TFBS | transcription factor binding sites |
| vg | viral genomes (aka gc, genome copies) |
| WPRE | Woodchuck hepatitis virus post-transcriptional regulatory element |
| wtAAV | Wild-type adeno-associated virus |
References
- Rodrigues, G.A.; Shalaev, E.; Karami, T.K.; Cunningham, J.; Slater, N.K.H.; Rivers, H.M. Pharmaceutical Development of AAV-Based Gene Therapy Products for the Eye. Pharm. Res. 2019, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boye, S.L.S.E.; Boye, S.L.S.E.; Lewin, A.S.; Hauswirth, W.W. A comprehensive review of retinal gene therapy. Mol. Ther. 2013, 21, 509–519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vandenberghe, L.H.; Wilson, J.M.; Gao, G. Tailoring the AAV vector capsid for gene therapy. Gene Ther. 2009, 16, 311–319. [Google Scholar] [CrossRef]
- Wang, D.; Tai, P.W.L.; Gao, G. Adeno-associated virus vector as a platform for gene therapy delivery. Nat. Rev. Drug Discov. 2019, 18, 358–378. [Google Scholar] [CrossRef]
- Cheever, T.R.; Berkley, D.; Braun, S.; Brown, R.H.; Byrne, B.J.; Chamberlain, J.S.; Cwik, V.; Duan, D.; Federoff, H.J.; High, K.A.; et al. Perspectives on Best Practices for Gene Therapy Programs. Hum. Gene Ther. 2015, 26, 127–133. [Google Scholar] [CrossRef]
- Lipinski, D.M.; Thake, M.; Maclaren, R.E. Clinical applications of retinal gene therapy. Prog. Retin. Eye Res. Clin. Appl. Retin. Gene Ther. 2013, 32, 22–47. [Google Scholar] [CrossRef]
- McClements, M.E.; MacLaren, R.E. Gene therapy for retinal disease. Transl. Res. 2013, 161, 241–254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, A.J.; Bainbridge, J.W.; Ali, R.R. Prospects for retinal gene replacement therapy. Trends Genet. 2009, 25, 156–165. [Google Scholar] [CrossRef]
- Bainbridge, J.W.B.; Tan, M.H.; Ali, R.R. Gene therapy progress and prospects: The eye. Gene Ther. 2006, 13, 1191–1197. [Google Scholar] [CrossRef]
- Duncan, J.L.; Pierce, E.A.; Laster, A.M.; Daiger, S.P.; Birch, D.G.; Ash, J.D.; Iannaccone, A.; Flannery, J.G.; Sahel, J.A.; Zack, D.J.; et al. Inherited retinal degenerations: Current landscape and knowledge gaps. Transl. Vis. Sci. Technol. 2018, 7. [Google Scholar] [CrossRef] [Green Version]
- Fahim, A.T.; Daiger, S.P.; Weleber, R.G. Nonsyndromic Retinitis Pigmentosa Overview. Available online: http://0-www-ncbi-nlm-nih-gov.brum.beds.ac.uk/pubmed/20301590 (accessed on 19 January 2017).
- Waehler, R.; Russell, S.J.; Curiel, D.T. Engineering targeted viral vectors for gene therapy. Nat. Rev. Genet. 2007, 8, 573–587. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Bao, J.; Zhang, Y.; Li, Z.; Zhou, X.; Wan, C.; Huang, L.; Zhao, Y.; Han, G.; Xue, T. Mammalian Near-Infrared Image Vision through Injectable and Self-Powered Retinal Nanoantennae. Cell 2019, 177, 243–255.e15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ziccardi, L.; Cordeddu, V.; Gaddini, L.; Matteucci, A.; Parravano, M.; Malchiodi-Albedi, F.; Varano, M. Gene therapy in retinal dystrophies. Int. J. Mol. Sci. 2019, 20, 5722. [Google Scholar] [CrossRef] [Green Version]
- Zallocchi, M.; Binley, K.; Lad, Y.; Ellis, S.; Widdowson, P.; Iqball, S.; Scripps, V.; Kelleher, M.; Loader, J.; Miskin, J.; et al. EIAV-based retinal gene therapy in the shaker1 mouse model for usher syndrome type 1B: Development of UshStat. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Truran, R.; Buckley, R.; Radcliffe, P.; Miskin, J.; Mitrophanous, K. Virus Purification. U.S. Patent 9169491B2, 27 October 2015. [Google Scholar]
- Payne, S.L.; Rausch, J.; Rushlow, K.; Montelaro, R.C.; Issel, C.; Flaherty, M.; Perry, S.; Sellon, D.; Fuller, F. Characterization of infectious molecular clones of equine infectious anaemia virus. J. Gen. Virol. 1994, 75, 425–429. [Google Scholar] [CrossRef]
- Kong, J.; Kim, S.-R.R.; Binley, K.; Pata, I.; Doi, K.; Mannik, J.; Zernant-Rajang, J.; Kan, O.; Iqball, S.; Naylor, S.; et al. Correction of the disease phenotype in the mouse model of Stargardt disease by lentiviral gene therapy. Gene Ther. 2008, 15, 1311–1320. [Google Scholar] [CrossRef] [Green Version]
- Vázquez-Domínguez, I.; Garanto, A.; Collin, R.W.J. Molecular Therapies for Inherited Retinal Diseases-Current Standing, Opportunities and Challenges. Genes 2019, 10, 654. [Google Scholar] [CrossRef] [Green Version]
- Aschauer, D.F.; Kreuz, S.; Rumpel, S. Analysis of Transduction Efficiency, Tropism and Axonal Transport of AAV Serotypes 1, 2, 5, 6, 8 and 9 in the Mouse Brain. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [Green Version]
- Duong, T.T.; Lim, J.; Vasireddy, V.; Papp, T.; Nguyen, H.; Leo, L.; Pan, J.; Zhou, S.; Chen, I.; Bennett, J.; et al. Comparative AAV-EGFP transgene expression using vector serotypes 1–9, 7M8, and 8b in human pluripotent stem cells, RPEs, and human and rat cortical neurons. Stem Cells Int. 2019, 2019. [Google Scholar] [CrossRef]
- Gardiner, K.L.; Cideciyan, A.V.; Swider, M.; Dufour, V.L.; Sumaroka, A.; Komáromy, A.M.; Hauswirth, W.W.; Iwabe, S.; Jacobson, S.G.; Beltran, W.A.; et al. Long-term Structural Outcomes of Late-stage RPE65 Gene Therapy. Mol. Ther. 2020, 28, 266–278. [Google Scholar] [CrossRef] [PubMed]
- Schnepp, B.C.; Clark, K.R.; Klemanski, D.L.; Pacak, C.A.; Johnson, P.R. Genetic Fate of Recombinant Adeno-Associated Virus Vector Genomes in Muscle. J. Virol. 2003, 77, 3495–3504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pellissier, L.P.; Hoek, R.M.; Vos, R.M.; Aartsen, W.M.; Klimczak, R.R.; Hoyng, S.A.; Flannery, J.G.; Wijnholds, J. Specific tools for targeting and expression in Müller glial cells. Mol. Ther.-Methods Clin. Dev. 2014, 1, 14009. [Google Scholar] [CrossRef] [PubMed]
- Koerber, J.T.; Klimczak, R.; Jang, J.-H.H.; Dalkara, D.; Flannery, J.G.; Schaffer, D.V. Molecular evolution of adeno-associated virus for enhanced glial gene delivery. Mol. Ther. 2009, 17, 2088–2095. [Google Scholar] [CrossRef]
- Pleticha, J.; Heilmann, L.F.; Evans, C.H.; Asokan, A.; Samulski, R.J.; Beutler, A.S. Preclinical toxicity evaluation of AAV for pain: Evidence from human AAV studies and from the pharmacology of analgesic drugs. Mol. Pain 2014, 10. [Google Scholar] [CrossRef] [Green Version]
- Calcedo, R.; Wilson, J.M. Humoral Immune Response to AAV. Front. Immunol. 2013, 4. [Google Scholar] [CrossRef] [Green Version]
- Tan, M.H.; Smith, A.J.; Pawlyk, B.; Xu, X.; Liu, X.; Bainbridge, J.B.; Basche, M.; McIntosh, J.; Tran, H.V.; Nathwani, A.; et al. Gene therapy for retinitis pigmentosa and Leber congenital amaurosis caused by defects in AIPL1: Effective rescue of mouse models of partial and complete Aipl1 deficiency using AAV2/2 and AAV2/8 vectors. Hum. Mol. Genet. 2009, 18, 2099–2114. [Google Scholar] [CrossRef]
- Francis, P.J. Genetics of inherited retinal disease. J. R. Soc. Med. 2006, 99, 189–191. [Google Scholar] [CrossRef] [Green Version]
- Den Hollander, A.I.; Black, A.; Bennett, J.; Cremers, F.P.M. Lighting a candle in the dark: Advances in genetics and gene therapy of recessive retinal dystrophies. J. Clin. Investig. 2010, 120, 3042–3053. [Google Scholar] [CrossRef] [Green Version]
- Daiger, S.; Sullivan, L.; Bowne, S. RetNet. Available online: https://sph.uth.edu/retnet/sum-dis.htm#A-genes (accessed on 11 June 2020).
- Schachat, A.P.; Maumenee, I.H. Bardet-Biedl Syndrome and Related Disorders. Arch. Ophthalmol. 1982, 100, 285–288. [Google Scholar] [CrossRef]
- Bocquet, B.; Lacroux, A.; Surget, M.-O.; Baudoin, C.; Marquette, V.; Manes, G.; Hebrard, M.; Sénéchal, A.; Delettre, C.; Roux, A.-F.; et al. Relative frequencies of inherited retinal dystrophies and optic neuropathies in Southern France: Assessment of 21-year data management. Ophthalmic Epidemiol. 2013, 20, 13–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lei, B.; Zhang, K.; Yue, Y.; Ghosh, A.; Duan, D. Retinal Degenerative Diseases; Spring: New York, NY, USA, 2010; Volume 664, pp. 671–678. [Google Scholar]
- Bainbridge, J.W.B.; Smith, A.J.; Barker, S.S.; Robbie, S.; Henderson, R.; Balaggan, K.; Viswanathan, A.; Holder, G.E.; Stockman, A.; Tyler, N.; et al. Effect of gene therapy on visual function in Leber’s congenital amaurosis. N. Engl. J. Med. 2008, 358, 2231–2239. [Google Scholar] [CrossRef] [PubMed]
- Georgiadis, A.; Duran, Y.; Ribeiro, J.; Abelleira-Hervas, L.; Robbie, S.J.; Sünkel-Laing, B.; Fourali, S.; Gonzalez-Cordero, A.; Cristante, E.; Michaelides, M.; et al. Development of an optimized AAV2/5 gene therapy vector for Leber congenital amaurosis owing to defects in RPE65. Gene Ther. 2016, 23, 857–862. [Google Scholar] [CrossRef]
- Samulski, R.J.; Berns, K.I.; Tan, M.; Muzyczka, N. Cloning of adeno-associated virus into pBR322: Rescue of intact virus from the recombinant plasmid in human cells. Proc. Natl. Acad. Sci. USA 1982, 79, 2077–2081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laughlin, C.A.; Tratschin, J.D.; Coon, H.; Carter, B.J. Cloning of infectious adeno-associated virus genomes in bacterial plasmids. Gene 1983, 23, 65–73. [Google Scholar] [CrossRef]
- Bennett, J. Gene Therapy for Leber’s Congenital Amaurosis Due to RPE65 Mutations. In Gene- and Cell-Based Treatment Strategies for the Eye; Rakoczy, E.P., Ed.; Springer: Berlin/Heidelberg, Germany, 2015; pp. 9–25. ISBN 978-3-662-45187-8. [Google Scholar]
- Yang, S.; Ma, S.Q.; Wan, X.; He, H.; Pei, H.; Zhao, M.J.; Chen, C.; Wang, D.W.; Dong, X.Y.; Yuan, J.J.; et al. Long-term outcomes of gene therapy for the treatment of Leber’s hereditary optic neuropathy. EBioMedicine 2016, 10, 258–268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grishanin, R.; Vuillemenot, B.; Sharma, P.; Keravala, A.; Greengard, J.; Gelfman, C.; Blumenkrantz, M.; Lawrence, M.; Hu, W.; Kiss, S.; et al. Preclinical Evaluation of ADVM-022, a Novel Gene Therapy Approach to Treating Wet Age-Related Macular Degeneration. Mol. Ther. 2019, 27, 118–129. [Google Scholar] [CrossRef] [Green Version]
- Ye, G.; Budzynski, E.; Sonnentag, P.; Nork, T.M.; Miller, P.E.; Sharma, A.K.; Ver Hoeve, J.N.; Smith, L.M.; Arndt, T.; Calcedo, R.; et al. Safety and Biodistribution Evaluation in Cynomolgus Macaques of rAAV2tYF-PR1.7-hCNGB3, a Recombinant AAV Vector for Treatment of Achromatopsia. Hum. Gene Ther. Clin. Dev. 2016, 27, 37–48. [Google Scholar] [CrossRef] [Green Version]
- Song, C.; Conlon, T.J.; Deng, W.T.; Coleman, K.E.; Zhu, P.; Plummer, C.; Mandapati, S.; Van Hoosear, M.; Green, K.B.; Sonnentag, P.; et al. Toxicology and pharmacology of an AAV vector expressing codon-optimized RPGR in RPGR-deficient Rd9 mice. Hum. Gene Ther. Clin. Dev. 2018, 29, 188–197. [Google Scholar] [CrossRef]
- Rumachik, N.G.; Malaker, S.A.; Poweleit, N.; Maynard, L.H.; Adams, C.M.; Leib, R.D.; Cirolia, G.; Thomas, D.; Stamnes, S.; Holt, K.; et al. Methods Matter—Standard Production Platforms For Recombinant AAV Can Produce Chemically And Functionally Distinct Vectors. bioRxiv 2019, 640169. [Google Scholar]
- Smale, S.T.; Kadonaga, J.T. The RNA Polymerase II Core Promoter. Annu. Rev. Biochem. 2003, 72, 449–479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chanda, D.; Hensel, J.A.; Higgs, J.T.; Grover, R.; Kaza, N.; Ponnazhagan, S. Effects of cellular methylation on transgene expression and site-specific integration of adeno-associated virus. Genes 2017, 8, 232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faust, S.M.; Bell, P.; Cutler, B.J.; Ashley, S.N.; Zhu, Y.; Rabinowitz, J.E.; Wilson, J.M. CpG-depleted adeno-associated virus vectors evade immune detection. J. Clin. Investig. 2013, 123, 2994–3001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Even, D.Y.; Kedmi, A.; Basch-Barzilay, S.; Ideses, D.; Tikotzki, R.; Shir-Shapira, H.; Shefi, O.; Juven-Gershon, T. Engineered promoters for potent transient overexpression. PLoS ONE 2016, 11. [Google Scholar] [CrossRef] [Green Version]
- Hartl, D.; Schübeler, D.; Roska, B.; Krebs, A.; Jüttner, J. Synp161, a Promoter for the Specific Expression of Genes in Rod Photoreceptors. WO2017093935A1, 8 June 2017. [Google Scholar]
- Jüttner, J.; Szabo, A.; Gross-Scherf, B.; Morikawa, R.K.; Rompani, S.B.; Hantz, P.; Szikra, T.; Esposti, F.; Cowan, C.S.; Bharioke, A.; et al. Targeting neuronal and glial cell types with synthetic promoter AAVs in mice, non-human primates and humans. Nat. Neurosci. 2019, 22, 1345–1356. [Google Scholar] [CrossRef]
- Xu, L.; Daly, T.; Gao, C.; Flotte, T.R.; Song, S.; Byrne, B.J.; Sands, M.S.; Parker Ponder, K. CMV-beta-actin promoter directs higher expression from an adeno-associated viral vector in the liver than the cytomegalovirus or elongation factor 1 alpha promoter and results in therapeutic levels of human factor X in mice. Hum. Gene Ther. 2001, 12, 563–573. [Google Scholar] [CrossRef] [Green Version]
- Miyazaki, M.; Ikeda, Y.; Yonemitsu, Y.; Goto, Y.; Sakamoto, T.; Tabata, T.; Ueda, Y.; Hasegawa, M.; Tobimatsu, S.; Ishibashi, T.; et al. Simian lentiviral vector-mediated retinal gene transfer of pigment epithelium-derived factor protects retinal degeneration and electrical defect in Royal College of Surgeons rats. Gene Ther. 2003, 10, 1503–1511. [Google Scholar] [CrossRef] [Green Version]
- Gilham, D.E.; Lie-A-Ling, M.; Taylor, N.; Hawkins, R.E. Cytokine stimulation and the choice of promoter are critical factors for the efficient transduction of mouse T cells with HIV-1 vectors. J. Gene Med. 2010, 12, 129–136. [Google Scholar] [CrossRef]
- Gray, S.J.; Foti, S.B.; Schwartz, J.W.; Bachaboina, L.; Taylor-Blake, B.; Coleman, J.; Ehlers, M.D.; Zylka, M.J.; McCown, T.J.; Samulski, R.J. Optimizing promoters for recombinant adeno-associated virus-mediated gene expression in the peripheral and central nervous system using self-complementary vectors. Hum. Gene Ther. 2011, 22, 1143–1153. [Google Scholar] [CrossRef] [Green Version]
- Ohlfest, J.R.; Frandsen, J.L.; Fritz, S.; Lobitz, P.D.; Perkinson, S.G.; Clark, K.J.; Nelsestuen, G.; Key, N.S.; McIvor, R.S.; Hackett, P.B.; et al. Phenotypic correction and long-term expression of factor VIII in hemophilic mice by immunotolerization and nonviral gene transfer using the Sleeping Beauty transposon system. Blood 2005, 105, 2691–2698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCown, T.J.; Xiao, X.; Li, J.; Breese, G.R.; Samulski, R.J. Differential and persistent expression patterns of CNS gene transfer by an adeno-associated virus (AAV) vector. Brain Res. 1996, 713, 99–107. [Google Scholar] [CrossRef]
- Powell, S.K.; Rivera-Soto, R.; Gray, S.J. Viral expression cassette elements to enhance transgene target specificity and expression in gene therapy. Discov. Med. 2015, 19, 49–57. [Google Scholar]
- Klein, R.L.; Meyer, E.M.; Peel, A.L.; Zolotukhin, S.; Meyers, C.; Muzyczka, N.; King, M.A. Neuron-specific transduction in the rat septohippocampal or nigrostriatal pathway by recombinant adeno-associated virus vectors. Exp. Neurol. 1998, 150, 183–194. [Google Scholar] [CrossRef]
- Farjo, R.; Skaggs, J.; Quiambao, A.B.; Cooper, M.J.; Naash, M.I. Efficient non-viral ocular gene transfer with compacted DNA nanoparticles. PLoS ONE 2006, 1. [Google Scholar] [CrossRef] [PubMed]
- Pellissier, L.P.; Quinn, P.M.; Henrique Alves, C.; Vos, R.M.; Klooster, J.; Flannery, J.G.; Alexander Heimel, J.; Wijnholds, J. Gene therapy into photoreceptors and Müller glial cells restores retinal structure and function in CRB1 retinitis pigmentosa mouse models. Hum. Mol. Genet. 2015, 24, 3104–3118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsai, Y.T.; Wu, W.H.; Lee, T.T.; Wu, W.P.; Xu, C.L.; Park, K.S.; Cui, X.; Justus, S.; Lin, C.S.; Jauregui, R.; et al. Clustered Regularly Interspaced Short Palindromic Repeats-Based Genome Surgery for the Treatment of Autosomal Dominant Retinitis Pigmentosa. Ophthalmology 2018, 125, 1421–1430. [Google Scholar] [CrossRef]
- Quinn, P.M.; Buck, T.M.; Mulder, A.A.; Ohonin, C.; Alves, C.H.; Vos, R.M.; Bialecka, M.; van Herwaarden, T.; van Dijk, E.H.C.C.; Talib, M.; et al. Human iPSC-Derived Retinas Recapitulate the Fetal CRB1 CRB2 Complex Formation and Demonstrate that Photoreceptors and Müller Glia Are Targets of AAV5. Stem Cell Rep. 2019, 12, 906–919. [Google Scholar] [CrossRef] [Green Version]
- Niwa, H.; Yamamura, K.; Miyazaki, J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 1991, 108, 193–199. [Google Scholar]
- Wang, Z.; Ma, H.-I.; Li, J.; Sun, L.; Zhang, J.; Xiao, X. Rapid and highly efficient transduction by double-stranded adeno-associated virus vectors in vitro and in vivo. Gene Ther. 2003, 10, 2105–2111. [Google Scholar] [CrossRef]
- Sawicki, J.A.; Morris, R.J.; Monks, B.; Sakai, K.; Miyazaki, J. SHORT NOTE A Composite CMV-IE Enhancer/β-Actin Promoter Is Ubiquitously Expressed in Mouse Cutaneous Epithelium. Exp. Cell Res. 1998, 244, 367–369. [Google Scholar] [CrossRef] [PubMed]
- Koilkonda, R.; Yu, H.; Talla, V.; Porciatti, V.; Feuer, W.J.; Hauswirth, W.W.; Chiodo, V.; Erger, K.E.; Boye, S.L.; Lewin, A.S.; et al. LHON gene therapy vector prevents visual loss and optic neuropathy induced by G11778A mutant mitochondrial DNA: Biodistribution and toxicology profile. Investig. Ophthalmol. Vis. Sci. 2014, 55, 7739–7753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bobo, R.H.; Laske, D.W.; Akbasak, A.; Morrison, P.F.; Dedrick, R.L.; Oldfield, E.H. Convection-enhanced delivery of macromolecules in the brain. Proc. Natl. Acad. Sci. USA 1994, 91, 2076–2080. [Google Scholar] [CrossRef] [Green Version]
- Rastegar, M.; Hotta, A.; Pasceri, P.; Makarem, M.; Cheung, A.Y.L.; Elliott, S.; Park, K.J.; Adachi, M.; Jones, F.S.; Clarke, I.D.; et al. MECP2 isoform-specific vectors with regulated expression for Rett syndrome gene therapy. PLoS ONE 2009, 4, e6810. [Google Scholar] [CrossRef]
- Li, C.; Hirsch, M.; Carter, P.; Asokan, A.; Zhou, X.; Wu, Z.; Samulski, R.J. A small regulatory element from chromosome 19 enhances liver-specific gene expression. Gene Ther. 2009, 16, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Gill, D.R.; Smyth, S.E.; Goddard, C.A.; Pringle, I.A.; Higgins, C.F.; Colledge, W.H.; Hyde, S.C. Increased persistence of lung gene expression using plasmids containing the ubiquitin C or elongation factor 1alpha promoter. Gene Ther. 2001, 8, 1539–1546. [Google Scholar] [CrossRef] [Green Version]
- Lu, Y.; Krishnan, A.; Brommer, B.; Tian, X.; Meer, M.; Vera, D.L.; Wang, C.; Zeng, Q.; Yu, D.; Bonkowski, M.S.; et al. Reversal of ageing- and injury-induced vision loss by Tet-dependent epigenetic reprogramming. bioRxiv 2019, 710210. [Google Scholar]
- Bochkov, Y.A.; Palmenberg, A.C. Translational efficiency of EMCV IRES in bicistronic vectors is dependent upon IRES sequence and gene location. Biotechniques 2006, 41, 283–292. [Google Scholar] [CrossRef] [Green Version]
- Attal, J.; Theron, M.C.; Puissant, C.; Houdebine, L.M. Effect of intercistronic length on internal ribosome entry site (IRES) efficiency in bicistronic mRNA. Gene Expr. 1999, 8, 299–309. [Google Scholar]
- Al-Allaf, F.A.; Abduljaleel, Z.; Athar, M.; Taher, M.M.; Khan, W.; Mehmet, H.; Colakogullari, M.; Apostolidou, S.; Bigger, B.; Waddington, S.; et al. Modifying inter-cistronic sequence significantly enhances IRES dependent second gene expression in bicistronic vector: Construction of optimised cassette for gene therapy of familial hypercholesterolemia. Non-Coding RNA Res. 2019, 4, 1–14. [Google Scholar] [CrossRef]
- Fagoe, N.D.; Eggers, R.; Verhaagen, J.; Mason, M.R.J. A compact dual promoter adeno-associated viral vector for efficient delivery of two genes to dorsal root ganglion neurons. Gene Ther. 2014, 21, 242–252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osakada, F.; Takahashi, M. Challenges in retinal circuit regeneration: Linking neuronal connectivity to circuit function. Biol. Pharm. Bull. 2015, 38, 341–357. [Google Scholar] [CrossRef] [Green Version]
- Tervo, D.G.R.; Hwang, B.Y.; Viswanathan, S.; Gaj, T.; Lavzin, M.; Ritola, K.D.; Lindo, S.; Michael, S.; Kuleshova, E.; Ojala, D.; et al. A Designer AAV Variant Permits Efficient Retrograde Access to Projection Neurons. Neuron 2016, 92, 372–382. [Google Scholar] [CrossRef] [Green Version]
- Dana, H.; Sun, Y.; Mohar, B.; Hulse, B.K.; Kerlin, A.M.; Hasseman, J.P.; Tsegaye, G.; Tsang, A.; Wong, A.; Patel, R.; et al. High-performance calcium sensors for imaging activity in neuronal populations and microcompartments. Nat. Methods 2019, 16, 649–657. [Google Scholar] [CrossRef]
- Nickells, R.W.; Schmitt, H.M.; Maes, M.E.; Schlamp, C.L. AAV2-mediated transduction of the mouse retina after optic nerve injury. Investig. Ophthalmol. Vis. Sci. 2017, 58, 6091–6104. [Google Scholar] [CrossRef] [PubMed]
- Garanto, A. RNA-Based Therapeutic Strategies for Inherited Retinal Dystrophies. Adv. Exp. Med. Biol. 2019, 1185, 71–77. [Google Scholar] [PubMed]
- Ren, R.; Li, Y.; Liu, Z.; Liu, K.; He, S. Long-term rescue of rat retinal ganglion cells and visual function by AAV-mediated BDNF expression after acute elevation of intraocular pressure. Investig. Ophthalmol. Vis. Sci. 2012, 53, 1003–1011. [Google Scholar] [CrossRef]
- LeVaillant, C.J.; Sharma, A.; Muhling, J.; Wheeler, L.P.; Cozens, G.S.; Hellström, M.; Rodger, J.; Harvey, A.R. Significant changes in endogenous retinal gene expression assessed 1 year after a single intraocular injection of AAV-CNTF or AAV-BDNF. Mol. Ther.-Methods Clin. Dev. 2016, 3, 16078. [Google Scholar] [CrossRef] [PubMed]
- Lau, D.; McGee, L.H.; Zhou, S.; Rendahl, K.G.; Manning, W.C.; Escobedo, J.A.; Flannery, J.G. Retinal degeneration is slowed in transgenic rats by AAV-mediated delivery of FGF-2. Investig. Ophthalmol. Vis. Sci. 2000, 41, 3622–3633. [Google Scholar]
- Leaver, S.G.; Cui, Q.; Plant, G.W.; Arulpragasam, A.; Hisheh, S.; Verhaagen, J.; Harvey, A.R. AAV-mediated expression of CNTF promotes long-term survival and regeneration of adult rat retinal ganglion cells. Gene Ther. 2006, 13, 1328–1341. [Google Scholar] [CrossRef] [Green Version]
- Dalkara, D.; Kolstad, K.D.; Guerin, K.I.; Hoffmann, N.V.; Visel, M.; Klimczak, R.R.; Schaffer, D.V.; Flannery, J.G. AAV mediated GDNF secretion from retinal glia slows down retinal degeneration in a rat model of retinitis pigmentosa. Mol. Ther. 2011, 19, 1602–1608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osborne, A.; Wang, A.X.Z.; Tassoni, A.; Widdowson, P.S.; Martin, K.R. Design of a Novel Gene Therapy Construct to Achieve Sustained Brain-Derived Neurotrophic Factor Signaling in Neurons. Hum. Gene Ther. 2018, 29, 828–841. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Askou, A.L.; Alsing, S.; Benckendorff, J.N.E.; Holmgaard, A.; Mikkelsen, J.G.; Aagaard, L.; Bek, T.; Corydon, T.J. Suppression of Choroidal Neovascularization by AAV-Based Dual-Acting Antiangiogenic Gene Therapy. Mol. Ther.-Nucleic Acids 2019, 16, 38–50. [Google Scholar] [CrossRef] [Green Version]
- Grimm, D.; Streetz, K.L.; Jopling, C.L.; Storm, T.A.; Pandey, K.; Davis, C.R.; Marion, P.; Salazar, F.; Kay, M.A. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature 2006, 441, 537–541. [Google Scholar] [CrossRef] [PubMed]
- Vázquez-Chona, F.R.; Clark, A.M.; Levine, E.M. Rlbp1 promoter drives robust Müller glial GFP expression in transgenic mice. Investig. Ophthalmol. Vis. Sci. 2009, 50, 3996–4003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanlon, K.S.; Chadderton, N.; Palfi, A.; Fernandez, A.B.; Humphries, P.; Kenna, P.F.; Millington-Ward, S.; Farrar, G.J. A novel retinal ganglion cell promoter for utility in AAV vectors. Front. Neurosci. 2017, 11. [Google Scholar] [CrossRef] [Green Version]
- Khani, S.C.; Pawlyk, B.S.; Bulgakov, O.V.; Kasperek, E.; Young, J.E.; Adamian, M.; Sun, X.; Smith, A.J.; Ali, R.R.; Li, T. AAV-mediated expression targeting of rod and cone photoreceptors with a human rhodopsin kinase promoter. Investig. Ophthalmol. Vis. Sci. 2007, 48, 3954–3961. [Google Scholar] [CrossRef] [Green Version]
- Isomura, H.; Stinski, M.F.; Kudoh, A.; Daikoku, T.; Shirata, N.; Tsurumi, T. Two Sp1/Sp3 Binding Sites in the Major Immediate-Early Proximal Enhancer of Human Cytomegalovirus Have a Significant Role in Viral Replication. J. Virol. 2005, 79, 9597–9607. [Google Scholar] [CrossRef] [Green Version]
- Dynan, W.S.; Tjian, R. The promoter-specific transcription factor Sp1 binds to upstream sequences in the SV40 early promoter. Cell 1983, 35, 79–87. [Google Scholar] [CrossRef]
- Briggs, M.R.; Kadonaga, J.T.; Bell, S.P.; Tjian, R. Purification and biochemical characterization of the promoter-specific transcription factor, Sp1. Science 1986, 234, 47–52. [Google Scholar] [CrossRef]
- Hebbard, L.; Steffen, A.; Zawadzki, V.; Fieber, C.; Howells, N.; Moll, J.; Ponta, H.; Hofmann, M.; Sleeman, J. CD44 expression and regulation during mammary gland development and function. J. Cell Sci. 2000, 113, 2619–2630. [Google Scholar] [PubMed]
- Su, M.; Hu, H.; Lee, Y.; D’Azzo, A.; Messing, A.; Brenner, M. Expression Specificity of GFAP Transgenes. Neurochem. Res. 2004, 29, 2075–2093. [Google Scholar] [CrossRef] [PubMed]
- Prentice, H.M.; Biswal, M.R.; Dorey, C.K.; Blanks, J.C. Hypoxia-regulated retinal glial cell-specific promoter for potential gene therapy in disease. Investig. Ophthalmol. Vis. Sci. 2011, 52, 8562–8570. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.S.; Matsuda, T.; Cepko, C.L. A core paired-type and POU homeodomain-containing transcription factor program drives retinal bipolar cell gene expression. J. Neurosci. 2008, 28, 7748–7764. [Google Scholar] [CrossRef] [Green Version]
- Aartsen, W.M.; van Cleef, K.W.R.R.; Pellissier, L.P.; Hoek, R.M.; Vos, R.M.; Blits, B.; Ehlert, E.M.E.E.; Balaggan, K.S.; Ali, R.R.; Verhaagen, J.; et al. GFAP-driven GFP expression in activated mouse Müller glial cells aligning retinal blood vessels following intravitreal injection of AAV2/6 vectors. PLoS ONE 2010, 5, e12387. [Google Scholar] [CrossRef]
- Dorrell, M.I.; Aguilar, E.; Jacobson, R.; Yanes, O.; Gariano, R.; Heckenlively, J.; Banin, E.; Ramirez, G.A.; Gasmi, M.; Bird, A.; et al. Antioxidant or neurotrophic factor treatment preserves function in a mouse model of neovascularization-associated oxidative stress. J. Clin. Investig. 2009, 119, 611–623. [Google Scholar] [CrossRef] [Green Version]
- Choi, V.; Bigelow, C.E.; Dryja, T.P.; Police, S.R. Viral Vectors for the Treatment of Retinal Dystrophy. U.S. Patent 9163259B2, 7 November 2013. [Google Scholar]
- Dougherty, C.J.; Smith, G.W.; Dorey, C.K.; Prentice, H.M.; Webster, K.A.; Blanks, J.C. Robust hypoxia-selective regulation of a retinal pigment epithelium-specific adeno-associated virus vector. Mol. Vis. 2008, 14, 471–480. [Google Scholar]
- Foster, L.C.; Wiesel, P.; Huggins, G.S.; Pañares, R.; Chin, M.T.; Pellacani, A.; Perrella, M.A. Role of activating protein-1 and high mobility group-I(Y) protein in the induction of CD44 gene expression by interleukin-1β in vascular smooth muscle cells. Faseb J. 2000, 14, 368–378. [Google Scholar] [CrossRef]
- Ichsan, A.M.; Kato, I.; Yoshida, T.; Takasawa, K.; Hayasaka, S.; Hiraga, K. Rhodopsin promoter-EGFP fusion transgene expression in photoreceptor neurons of retina and pineal complex in mice. Neurosci. Lett. 2005, 379, 138–143. [Google Scholar] [CrossRef] [Green Version]
- Allocca, M.; Mussolino, C.; Garcia-Hoyos, M.; Sanges, D.; Iodice, C.; Petrillo, M.; Vandenberghe, L.H.; Wilson, J.M.; Marigo, V.; Surace, E.M.; et al. Novel adeno-associated virus serotypes efficiently transduce murine photoreceptors. J. Virol. 2007, 81, 11372–11380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Komáromy, A.M.; Alexander, J.J.; Cooper, A.E.; Chiodo, V.A.; Glushakova, L.G.; Acland, G.M.; Hauswirth, W.W.; Aguirre, G.D. Targeting gene expression to cones with human cone opsin promoters in recombinant AAV. Gene Ther. 2008, 15, 1049–1055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quiambao, A.B.; Peachey, N.S.; Mangini, N.J.; Röhlich, P.; Hollyfield, J.G.; Al-Ubaidi, M.R. A 221-bp fragment of the mouse opsin promoter directs expression specifically to the rod photoreceptors of transgenic mice. Vis. Neurosci. 1997, 14, 617–625. [Google Scholar] [CrossRef] [PubMed]
- Young, J.E. A Short, Highly Active Photoreceptor-Specific Enhancer/Promoter Region Upstream of the Human Rhodopsin Kinase Gene. Investig. Ophthalmol. Vis. Sci. 2003, 44, 4076–4085. [Google Scholar] [CrossRef]
- Sun, X.; Pawlyk, B.; Xu, X.; Liu, X.; Bulgakov, O.V.; Adamian, M.; Sandberg, M.A.; Khani, S.C.; Tan, M.-H.; Smith, A.J.; et al. Gene therapy with a promoter targeting both rods and cones rescues retinal degeneration caused by AIPL1 mutations. Gene Ther. 2010, 17, 117–131. [Google Scholar] [CrossRef] [Green Version]
- Beltran, W.A.; Cideciyan, A.V.; Boye, S.E.; Ye, G.-J.; Iwabe, S.; Dufour, V.L.; Marinho, L.F.; Swider, M.; Kosyk, M.S.; Sha, J.; et al. Optimization of Retinal Gene Therapy for X-Linked Retinitis Pigmentosa Due to RPGR Mutations. Mol. Ther. 2017, 25, 1866–1880. [Google Scholar] [CrossRef]
- Pang, J.; Deng, W.-T.; Dai, X.; Lei, B.; Everhart, D.; Umino, Y.; Li, J.; Zhang, K.; Mao, S.; Boye, S.L.; et al. AAV-mediated cone rescue in a naturally occurring mouse model of CNGA3-achromatopsia. PLoS ONE 2012, 7, e35250. [Google Scholar] [CrossRef] [Green Version]
- Glushakova, L.G.; Timmers, A.M.; Pang, J.; Teusner, J.T.; Hauswirth, W.W. Human blue-opsin promoter preferentially targets reporter gene expression to rat s-cone photoreceptors. Investig. Ophthalmol. Vis. Sci. 2006, 47, 3505–3513. [Google Scholar] [CrossRef] [Green Version]
- Michalakis, S.; Mühlfriedel, R.; Tanimoto, N.; Krishnamoorthy, V.; Koch, S.; Fischer, M.D.; Becirovic, E.; Bai, L.; Huber, G.; Beck, S.C.; et al. Restoration of cone vision in the CNGA3-/- mouse model of congenital complete lack of cone photoreceptor function. Mol. Ther. 2010, 18, 2057–2063. [Google Scholar] [CrossRef]
- Beltran, W.A.; Cideciyan, A.V.; Lewin, A.S.; Iwabe, S.; Khanna, H.; Sumaroka, A. Gene therapy rescues photoreceptor blindness in dogs and paves the way for treating human X-linked retinitis pigmentosa. Proc. Natl. Acad. Sci. USA 2012, 106, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Dyka, F.; Boye, S.; Ryals, R.; Chiodo, V.; Boye, S.; Hauswirth, W. Cone specific promoter for use in gene therapy of retinal degenerative diseases. Adv Exp Med Biol 2014, 801, 695–701. [Google Scholar] [PubMed] [Green Version]
- Fischer, M.D.; Michalakis, S.; Wilhelm, B.; Zobor, D.; Muehlfriedel, R.; Kohl, S.; Weisschuh, N.; Ochakovski, G.A.; Klein, R.; Schoen, C.; et al. Safety and Vision Outcomes of Subretinal Gene Therapy Targeting Cone Photoreceptors in Achromatopsia: A Nonrandomized Controlled Trial [published online ahead of print, 30 April 2020]. JAMA Ophthalmol. 2020, 138, 643–651. [Google Scholar] [CrossRef] [PubMed]
- Pickrell, S.W.; Zhu, X.; Wang, X.; Craft, C.M. Deciphering the contribution of known cis-elements in the mouse cone arrestin gene to its cone-specific expression. Investig. Ophthalmol. Vis. Sci. 2004, 45, 3877–3884. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Timmers, A.M.; Guy, J.; Pang, J.; Hauswirth, W.W. Cone-specific expression using a human red opsin promoter in recombinant AAV. Vis. Res. 2008, 48, 332–338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Georgiadis, A.; Matsuki, T.; Rizzi, M.; Hoke, J.; Gonzalez-Cordero, A.; Sampson, R.; Bainbridge, J.; Smith, A.; Ali, R. ARVO Annual Meeting Poster A197 Development and efficacy assessment of AAV2/8-hG1.7p.coCNGA3, a CNGA3 gene therapy vector. Investig. Ophthalmol. Vis. Sci. 2019, 60, 3426. [Google Scholar]
- Rizzi, M.; Ali, R.; Smith, A.; Nishiguchi, K. Gene Therapy to Improve Vision. U.S. Patent 20180030477A1, 1 February 2018. [Google Scholar]
- Forbes, A. United States Securities and Exchange Commission (SEC) Form S-1 Registration Statement MeiraGTx Holdings plc; United States Securities and Exchange Commission: Washington, DC, USA, 2018. [Google Scholar]
- Watanabe, S.; Sanuki, R.; Ueno, S.; Koyasu, T.; Hasegawa, T.; Furukawa, T. Tropisms of AAV for Subretinal Delivery to the Neonatal Mouse Retina and Its Application for In Vivo Rescue of Developmental Photoreceptor Disorders. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [Green Version]
- Horsager, A.; Smith, A.; Matteo, B.C. Modulation Neural Pathways. WO 2012/051599 A2, 19 April 2012. [Google Scholar]
- Lu, Q.; Ganjawala, T.H.; Ivanova, E.; Cheng, J.G.; Troilo, D.; Pan, Z.H. AAV-mediated transduction and targeting of retinal bipolar cells with improved mGluR6 promoters in rodents and primates. Gene Ther. 2016, 23, 680–689. [Google Scholar] [CrossRef] [Green Version]
- Glover, C.P.J.; Bienemann, A.S.; Heywood, D.J.; Cosgrave, A.S.; Uney, J.B. Adenoviral-mediated, high-level, cell-specific transgene expression: A SYN1-WPRE cassette mediates increased transgene expression with no loss of neuron specificity. Mol. Ther. 2002, 5, 509–516. [Google Scholar] [CrossRef]
- Dalkara, D.; Picaud, S.; Desrosiers, M.; Sahel, J.-A.; Duebel, J.; Bemelmans, A.; Roska, B. Promoters and uses thereof. U.S. Patent 20180355354A1, 13 December 2018. [Google Scholar]
- Simpson, E.M.; Korecki, A.J.; Fornes, O.; McGill, T.J.; Cueva-Vargas, J.L.; Agostinone, J.; Farkas, R.A.; Hickmott, J.W.; Lam, S.L.; Mathelier, A.; et al. New MiniPromoter Ple345 (NEFL) drives strong and specific expression in retinal ganglion cells of mouse and primate retina. Hum. Gene Ther. 2019, 30, 257–272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali, R.R.; Bainbridge, J.W.B.; Smith, A.J. Devices and Methods for Delivering Polynucleotides into Retinal Cells of the Macula and Fovea. U.S. Patent 20100081707A1, 1 April 2010. [Google Scholar]
- Maddalena, A.; Tornabene, P.; Tiberi, P.; Minopoli, R.; Manfredi, A.; Mutarelli, M.; Rossi, S.; Simonelli, F.; Naggert, J.K.; Cacchiarelli, D.; et al. Triple Vectors Expand AAV Transfer Capacity in the Retina. Mol. Ther. 2018, 26, 524–541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jüttner, J.; Krol, J.; Roska, B. Synpiii, a Promoter for the Specific Expression of Genes in Retinal Pigment Epithelium. WO2019106035A1, 18 August 2016. [Google Scholar]
- Mäkinen, P.I.; Koponen, J.K.; Kärkkäinen, A.M.; Malm, T.M.; Pulkkinen, K.H.; Koistinaho, J.; Turunen, M.P.; Ylä-Herttuala, S. Stable RNA interference: Comparison of U6 and H1 promoters in endothelial cells and in mouse brain. J. Gene Med. 2006, 8, 433–441. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Harwig, A.; Berkhout, B.; Herrera-Carrillo, E. Mutation of nucleotides around the +1 position of type 3 polymerase III promoters: The effect on transcriptional activity and start site usage. Transcription 2017, 8, 275–287. [Google Scholar] [CrossRef] [Green Version]
- Ong, S.T.; Li, F.; Du, J.; Tan, Y.W.; Wang, S. Hybrid cytomegalovirus enhancer-h1 promoter-based plasmid and baculovirus vectors mediate effective RNA interference. Hum. Gene Ther. 2005, 16, 1404–1412. [Google Scholar] [CrossRef]
- Eckenfelder, A.; Tordo, J.; Babbs, A.; Davies, K.E.; Goyenvalle, A.; Danos, O. The Cellular Processing Capacity Limits the Amounts of Chimeric U7 snRNA Available for Antisense Delivery. Mol. Ther. Nucleic Acids 2012, 1, e31. [Google Scholar] [CrossRef]
- Salva, M.Z.; Himeda, C.L.; Tai, P.W.; Nishiuchi, E.; Gregorevic, P.; Allen, J.M.; Finn, E.E.; Nguyen, Q.G.; Blankinship, M.J.; Meuse, L.; et al. Design of tissue-specific regulatory cassettes for high-level rAAV-mediated expression in skeletal and cardiac muscle. Mol. Ther. 2007, 15, 320–329. [Google Scholar] [CrossRef]
- LaVail, M.M.; Yasumura, D.; Matthes, M.T.; Drenser, K.A.; Flannery, J.G.; Lewin, A.S.; Hauswirth, W.W. Ribozyme rescue of photoreceptor cells in P23H transgenic rats: Long-term survival and late-stage therapy. Proc. Natl. Acad. Sci. USA 2000, 97, 11488–11493. [Google Scholar]
- Xu, L.; Zhao, L.; Gao, Y.; Xu, J.; Han, R. Empower multiplex cell and tissue-specific CRISPR-mediated gene manipulation with self-cleaving ribozymes and tRNA. Nucleic Acids Res. 2017, 45. [Google Scholar] [CrossRef]
- Patrício, M.I.; Barnard, A.R.; Orlans, H.O.; McClements, M.E.; MacLaren, R.E. Inclusion of the Woodchuck Hepatitis Virus Posttranscriptional Regulatory Element Enhances AAV2-Driven Transduction of Mouse and Human Retina. Mol. Ther.-Nucleic Acids 2017, 6, 198–208. [Google Scholar]
- Choi, J.-H.; Yu, N.-K.; Baek, G.-C.; Bakes, J.; Seo, D.; Nam, H.J.; Baek, S.H.; Lim, C.-S.; Lee, Y.-S.; Kaang, B.-K. Optimization of AAV expression cassettes to improve packaging capacity and transgene expression iChoi J-H, Yu N-K, Baek G-C, Bakes J, Seo D, Nam HJ, Baek SH, Lim C-S, Lee Y-S, Kaang B-K. 2014. Optimization of AAV expression cassettes to improve packaging. Mol. Brain 2014, 7, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michalakis, S.; Biel, M.; Seeliger, M.; Schoen, C. Gene Therapy for the Treatment of a Retinal Degeneration Disease. U.S. Patent 20180353620A1, 30 August 2017. [Google Scholar]
- Ramezani, A.; Hawley, T.S.; Hawley, R.G. Lentiviral vectors for enhanced gene expression in human hematopoietic cells. Mol. Ther. 2000, 2, 458–469. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.; Yue, Y.; Liu, M.; Duan, D. Synthetic intron improves transduction efficiency of trans-splicing adeno-associated viral vectors. Hum. Gene Ther. 2006, 17, 1036–1042. [Google Scholar] [CrossRef] [Green Version]
- Wu, Z.; Sun, J.; Zhang, T.; Yin, C.; Yin, F.; Van Dyke, T.; Samulski, R.J.; Monahan, P.E. Optimization of self-complementary AAV vectors for liver-directed expression results in sustained correction of hemophilia B at low vector dose. Mol. Ther. 2008, 16, 280–289. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Williams, J.A.; Luke, J.; Zhang, F.; Chu, K.; Kay, M.A. A 5′ noncoding exon containing engineered intron enhances transgene expression from recombinant AAV vectors in vivo. Hum. Gene Ther. 2017, 28, 125–134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karali, M.; Manfredi, A.; Puppo, A.; Marrocco, E.; Gargiulo, A.; Allocca, M.; de Corte, M.; Rossi, S.; Giunti, M.; Bacci, M.L.; et al. MicroRNA-Restricted transgene expression in the retina. PLoS ONE 2011, 6. [Google Scholar] [CrossRef]
- Brown, B.D.; Cantore, A.; Annoni, A.; Sergi, L.S.; Lombardo, A.; Della Valle, P.; D’Angelo, A.; Naldini, L. A microRNA-regulated lentiviral vector mediates stable correction of hemophilia B mice. Blood 2007, 110, 4144–4152. [Google Scholar] [CrossRef] [Green Version]
- Xie, J.; Mao, Q.; Tai, P.W.L.; He, R.; Ai, J.; Su, Q.; Zhu, Y.; Ma, H.; Li, J.; Gong, S.; et al. Short DNA Hairpins Compromise Recombinant Adeno-Associated Virus Genome Homogeneity. Mol. Ther. 2017, 25, 1363–1374. [Google Scholar] [CrossRef] [Green Version]
- Domenger, C.; Grimm, D. Next-generation AAV vectors—Do not judge a virus (only) by its cover. Hum. Mol. Genet. 2019, 28, R3–R14. [Google Scholar] [CrossRef] [Green Version]
- Yew, N.S.; Wysokenski, D.M.; Wang, K.X.; Ziegler, R.J.; Marshall, J.; McNeilly, D.; Cherry, M.; Osburn, W.; Cheng, S.H. Optimization of plasmid vectors for high-level expression in lung epithelial cells. Hum. Mol. Genet. 1997, 8, 575–584. [Google Scholar] [CrossRef] [PubMed]
- Donello, J.E.; Loeb, J.E.; Hope, T.J. Woodchuck hepatitis virus contains a tripartite posttranscriptional regulatory element. J. Virol. 1998, 72, 5085–5092. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ronzitti, G.; Collaud, F.; Bortolussi, G.; Charles, S.; Vidal, P.; Sola, M.S.; Muro, A.; Mingozzi, F. 158. Cryptic ATG Removal from Synthetic Introns Increase the Therapeutic Efficacy of AAV Vector Mediated Gene Transfer. Mol. Ther. 2016, 24, S62. [Google Scholar] [CrossRef]
- Choi, V.W.; Bigelow, C.E.; McGee, T.L.; Gujar, A.N.; Li, H.; Hanks, S.M.; Vrouvlianis, J.; Maker, M.; Leehy, B.; Zhang, Y.; et al. AAV-mediated RLBP1 gene therapy improves the rate of dark adaptation in Rlbp1 knockout mice. Mol. Ther.-Methods Clin. Dev. 2015, 2, 15022. [Google Scholar] [CrossRef] [PubMed]
- Beltran, W.A.; Aguirre, G.D.; Jacobson, S.G.; Cideciyan, A.V.; Lewin, A.S.; Boye, S.L.; Hauswirth, W.W.; Deng, W.-T. AAV-Mediated Gene Therapy for RPGR X-Linked Retinal Degeneration. U.S. Patent 9770491B2, 26 September 2017. [Google Scholar]
- Xu, D.H.; Wang, X.Y.; Jia, Y.L.; Wang, T.Y.; Tian, Z.W.; Feng, X.; Zhang, Y.N. SV40 intron, a potent strong intron element that effectively increases transgene expression in transfected Chinese hamster ovary cells. J. Cell. Mol. Med. 2018, 22, 2231–2239. [Google Scholar] [CrossRef] [Green Version]
- MacLaren, R.E.; Groppe, M.; Barnard, A.R.; Cottriall, C.L.; Tolmachova, T.; Seymour, L.; Reed Clark, K.; During, M.J.; Cremers, F.P.M.; Black, G.C.M.; et al. Retinal gene therapy in patients with choroideremia: Initial fi ndings from a phase 1/2 clinical trial. Lancet 2014, 383, 1129–1137. [Google Scholar] [CrossRef] [Green Version]
- Jacobson, S.G.; Cideciyan, A.V.; Ratnakaram, R.; Heon, E.; Schwartz, S.B.; Roman, A.J.; Peden, M.C.; Aleman, T.S.; Boye, S.L.; Sumaroka, A.; et al. Gene therapy for leber congenital amaurosis caused by RPE65 mutations: Safety and efficacy in 15 children and adults followed up to 3 years. Arch. Ophthalmol. 2012, 130, 9–24. [Google Scholar] [CrossRef] [Green Version]
- Lam, B.L.; Davis, J.L.; Gregori, N.Z.; MacLaren, R.E.; Girach, A.; Verriotto, J.D.; Rodriguez, B.; Rosa, P.R.; Zhang, X.; Feuer, W.J. Choroideremia Gene Therapy Phase 2 Clinical Trial: 24-Month Results. Am. J. Ophthalmol. 2019, 197, 65–73. [Google Scholar] [CrossRef]
- Kurachi, S.; Hitomi, Y.; Furukawa, M.; Kurachi, K. Role of intron I in expression of the human factor IX gene. J. Biol. Chem. 1995, 270, 5276–5281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.H.; Lee, S.R.; Li, L.H.; Park, H.J.; Park, J.H.; Lee, K.Y.; Kim, M.K.; Shin, B.A.; Choi, S.Y. High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS ONE 2011, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Urabe, M.; Hasumi, Y.; Ogasawara, Y.; Matsushita, T.; Kamoshita, N.; Nomoto, A.; Colosi, P.; Kurtzman, G.J.; Tobita, K.; Ozawa, K. A novel dicistronic AAV vector using a short IRES segment derived from hepatitis C virus genome. Gene 1997, 200, 157–162. [Google Scholar] [CrossRef]
- Eggermont, J.; Proudfoot, N.J. Poly(A) signals and transcriptional pause sites combine to prevent interference between RNA polymerase II promoters. Embo J. 1993, 12, 2539–2548. [Google Scholar] [CrossRef]
- Mhyre, A.J.; Marcondes, A.M.; Spaulding, E.Y.; Deeg, H.J. Stroma-dependent apoptosis in clonal hematopoietic precursors correlates with expression of PYCARD. Blood 2009, 113, 649–658. [Google Scholar] [CrossRef] [Green Version]
- Satkunanathan, S.; Wheeler, J.; Thorpe, R.; Zhao, Y. Establishment of a novel cell line for the enhanced production of recombinant adeno-associated virus vectors for gene therapy. Hum. Gene Ther. 2014, 25, 929–941. [Google Scholar] [CrossRef] [Green Version]
- VectorBuilder Inc. Vectorbuilder.com/Learning-Center/Vector-Component/Promoter. Available online: https://en.vectorbuilder.com/learning-center/vector-component/linker.html (accessed on 11 June 2020).
- Martinez-Lopez, A.; Encinas, P.; García-Valtanen, P.; Gomez-Casado, E.; Coll, J.M.; Estepa, A. Improving the safety of viral DNA vaccines: Development of vectors containing both 5′ and 3′ homologous regulatory sequences from non-viral origin. Appl. Microbiol. Biotechnol. 2013, 97, 3007–3016. [Google Scholar] [CrossRef]
- McFarland, T.J.; Zhang, Y.; Atchaneeyaskul, L.O.; Francis, P.; Stout, J.T.; Appukuttan, B. Evaluation of a novel short polyadenylation signal as an alternative to the SV40 polyadenylation signal. Plasmid 2006, 56, 62–67. [Google Scholar] [CrossRef]
- Vora, S.; Cheng, J.; Xiao, R.; VanDusen, N.J.; Quintino, L.; Pu, W.T.; Vandenberghe, L.H.; Chavez, A.; Church, G. Rational design of a compact CRISPR-Cas9 activator for AAV-mediated delivery. bioRxiv 2018, 9, 298620. [Google Scholar]
- Hager, S.; Frame, F.M.; Collins, A.T.; Burns, J.E.; Maitland, N.J. An internal polyadenylation signal substantially increases expression levels of lentivirus-delivered transgenes but has the potential to reduce viral titer in a promoter-dependent manner. Hum. Gene Ther. 2008, 19, 840–850. [Google Scholar] [CrossRef]
- Schambach, A.; Galla, M.; Maetzig, T.; Loew, R.; Baum, C. Improving transcriptional termination of self-inactivating gamma-retroviral and lentiviral vectors. Mol. Ther. 2007, 15, 1167–1173. [Google Scholar] [CrossRef]
- Ostedgaard, L.S.; Rokhlina, T.; Karp, P.H.; Lashmit, P.; Afione, S.; Schmidt, M.; Zabner, J.; Stinski, M.F.; Chiorini, J.A.; Welsh, M.J. A shortened adeno-associated virus expression cassette for CFTR gene transfer to cystic fibrosis airway epithelia. Proc. Natl. Acad. Sci. USA 2005, 102, 2952–2957. [Google Scholar] [CrossRef] [Green Version]
- Kumar-Singh, R.; Leaderer, D.; Cashman, S. Compositions, Kits and Methods for Treatment of Complement-Related Disorders. U.S. Patent 20170209535A1, 16 July 2019. [Google Scholar]
- Cole, C.N.; Stacy, T.P. Identification of sequences in the herpes simplex virus thymidine kinase gene required for efficient processing and polyadenylation. Mol. Cell. Biol. 1985, 5, 2104–2113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lipinski, D.M. A Comparison of Inducible Gene Expression Platforms: Implications for Recombinant Adeno-Associated Virus (rAAV) Vector-Mediated Ocular Gene Therapy. In Advances in Experimental Medicine and Biology; Bowes Rickman, C., Grimm, C., Anderson, R., Ash, J., LaVail, M., Hollyfield, J.G., Eds.; Springer: Cham, Switzerland, 2019; Volume 1185, pp. 79–83. [Google Scholar]
- Peterson, M.G.; Mercer, J.F.B. Structure and regulation of the sheep metallothionein-Ia gene. Eur. J. Biochem. 1986, 160, 579–585. [Google Scholar] [CrossRef] [PubMed]
- Grimm, S.L.; Nordeen, S.K. Mouse mammary tumor virus sequences responsible for activating cellular oncogenes. J. Virol. 1998, 72, 9428–9435. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, K.R. Steroid Receptor Regulated Transcription of Specific Genes and Gene Networks. Annu. Rev. Genet. 1985, 19, 209–252. [Google Scholar] [CrossRef] [PubMed]
- Naughton, B.J.; Han, D.D.; Gu, H.H. Fluorescence-based evaluation of shRNA efficacy. Anal. Biochem. 2011, 417, 162–164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strobel, B.; Spöring, M.; Klein, H.; Blazevic, D.; Rust, W.; Sayols, S.; Hartig, J.S.; Kreuz, S. High-throughput identification of synthetic riboswitches by barcode-free amplicon-sequencing in human cells. Nat. Commun. 2020, 11, 714. [Google Scholar] [CrossRef]
- Reid, C.A.; Nettesheim, E.R.; Connor, T.B.; Lipinski, D.M. Development of an inducible anti-VEGF rAAV gene therapy strategy for the treatment of wet AMD. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef] [Green Version]
- Peränen, J.; Rikkonen, M.; Hyvönen, M.; Kääriäinen, L. T7 vectors with modified T7lac promoter for expression of proteins in Escherichia coli. Anal. Biochem. 1996, 236, 371–373. [Google Scholar] [CrossRef] [PubMed]
- Cenik, C.; Chua, H.N.; Zhang, H.; Tarnawsky, S.P.; Akef, A.; Derti, A.; Tasan, M.; Moore, M.J.; Palazzo, A.F.; Roth, F.P. Genome analysis reveals interplay between 5′UTR introns and nuclear mRNA export for secretory and mitochondrial genes. PLoS Genet. 2011, 7, e1001366. [Google Scholar] [CrossRef] [Green Version]
- Bonnet, A.; Grosso, A.R.; Elkaoutari, A.; Coleno, E.; Presle, A.; Sridhara, S.C.; Janbon, G.; Géli, V.; de Almeida, S.F.; Palancade, B. Introns Protect Eukaryotic Genomes from Transcription-Associated Genetic Instability. Mol. Cell 2017, 67, 608–621.e6. [Google Scholar] [CrossRef] [PubMed]
- Duan, D. Systemic AAV Micro-dystrophin Gene Therapy for Duchenne Muscular Dystrophy. Mol. Ther. 2018, 26, 2337–2356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- England, S.B.; Nicholson, L.V.; Johnson, M.A.; Forrest, S.M.; Love, D.R.; Zubrzycka-Gaarn, E.E.; Bulman, D.E.; Harris, J.B.; Davies, K.E. Very mild muscular dystrophy associated with the deletion of 46% of dystrophin. Nature 1990, 343, 180–182. [Google Scholar] [CrossRef]
- Davies, K.E.; Chamberlain, J.S. Surrogate gene therapy for muscular dystrophy. Nat. Med. 2019, 25, 1473–1474. [Google Scholar] [CrossRef]
- Zhang, W.; Li, L.; Su, Q.; Gao, G.; Khanna, H. Gene Therapy Using a miniCEP290 Fragment Delays Photoreceptor Degeneration in a Mouse Model of Leber Congenital Amaurosis. Hum. Gene Ther. 2018, 29, 42–50. [Google Scholar] [CrossRef] [Green Version]
- Dona, M.A. Gene therapy using miniUSH2A genes improves visual function in a zebrafish ush2a knock-out model. In Towards gene therapy for USH2A-associated retinitis pigmentosa. ~Fishing for answers~ [Doctoral Thesis]; Kremer, J.M.J., Keunen, J.E.E., van Wijk, H.A.R., Eds.; Radboud University Nijmegen: Nijmegen, The Netherlands, 2018; pp. 1–243. ISBN 978-94-6375-060-8. [Google Scholar]
- Song, Y.; Morales, L.; Malik, A.S.; Mead, A.F.; Greer, C.D.; Mitchell, M.A.; Petrov, M.T.; Su, L.T.; Choi, M.E.; Rosenblum, S.T.; et al. Non-immunogenic utrophin gene therapy for the treatment of muscular dystrophy animal models. Nat. Med. 2019, 25, 1505–1511. [Google Scholar] [CrossRef]
- Van Rossum, A.G.S.H.; Aartsen, W.M.; Meuleman, J.; Klooster, J.; Malysheva, A.; Versteeg, I.; Arsanto, J.P.; Le Bivic, A.; Wijnholds, J. Pals1/Mpp5 is required for correct localization of Crb1 at the subapical region in polarized Müller glia cells. Hum. Mol. Genet. 2006, 15, 2659–2672. [Google Scholar] [CrossRef]
- Alves, C.H.; Sanz, A.S.; Park, B.; Pellissier, L.P.; Tanimoto, N.; Beck, S.C.; Huber, G.; Murtaza, M.; Richard, F.; Sridevi Gurubaran, I.; et al. Loss of CRB2 in the mouse retina mimics human retinitis pigmentosa due to mutations in the CRB1 gene. Hum. Mol. Genet. 2013, 22, 35–50. [Google Scholar] [CrossRef] [Green Version]
- Alves, C.H.; Boon, N.; Mulder, A.A.; Koster, A.J.; Jost, C.R.; Wijnholds, J. CRB2 Loss in Rod Photoreceptors Is Associated with Progressive Loss of Retinal Contrast Sensitivity. Int. J. Mol. Sci. 2019, 20, 4069. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van de Pavert, S.A.; Meuleman, J.; Malysheva, A.; Aartsen, W.M.; Versteeg, I.; Tonagel, F.; Kamphuis, W.; McCabe, C.J.; Seeliger, M.W.; Wijnholds, J. A single amino acid substitution (Cys249Trp) in Crb1 causes retinal degeneration and deregulates expression of pituitary tumor transforming gene Pttg1. J. Neurosci. 2007, 27, 564–573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alves, C.H.; Bossers, K.; Vos, R.M.; Essing, A.H.W.; Swagemakers, S.; van der Spek, P.J.; Verhaagen, J.; Wijnholds, J. Microarray and morphological analysis of early postnatal CRB2 mutant retinas on a pure C57BL/6J genetic background. PLoS ONE 2013, 8, e82532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alves, C.H.; Pellissier, L.P.; Vos, R.M.; Garcia Garrido, M.; Sothilingam, V.; Seide, C.; Beck, S.C.; Klooster, J.; Furukawa, T.; Flannery, J.G.; et al. Targeted ablation of Crb2 in photoreceptor cells induces retinitis pigmentosa. Hum. Mol. Genet. 2014, 23, 3384–3401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quinn, P.M.; Alves, C.H.; Klooster, J.; Wijnholds, J. CRB2 in immature photoreceptors determines the superior-inferior symmetry of the developing retina to maintain retinal structure and function. Hum. Mol. Genet. 2018, 27, 3137–3153. [Google Scholar] [CrossRef] [Green Version]
- Van De Pavert, S.A.; Sanz, A.S.; Aartsen, W.M.; Vos, R.M.; Versteeg, I.; Beck, S.C.; Klooster, J.; Seeliger, M.W.; Wijnholds, J. Crb1 is a determinant of retinal apical Müller glia cell features. Glia 2007, 55, 1486–1497. [Google Scholar] [CrossRef]
- Van de Pavert, S.A.; Kantardzhieva, A.; Malysheva, A.; Meuleman, J.; Versteeg, I.; Levelt, C.; Klooster, J.; Geiger, S.; Seeliger, M.W.; Rashbass, P.; et al. Crumbs homologue 1 is required for maintenance of photoreceptor cell polarization and adhesion during light exposure. J. Cell Sci. 2004, 117, 4169–4177. [Google Scholar] [CrossRef] [Green Version]
- Pellissier, L.P.; Alves, C.H.; Quinn, P.M.; Vos, R.M.; Tanimoto, N.; Lundvig, D.M.S.; Dudok, J.J.; Hooibrink, B.; Richard, F.; Beck, S.C.; et al. Targeted ablation of CRB1 and CRB2 in retinal progenitor cells mimics Leber congenital amaurosis. PLoS Genet. 2013, 9, e1003976. [Google Scholar] [CrossRef]
- Pellissier, L.P.; Lundvig, D.M.S.; Tanimoto, N.; Klooster, J.; Vos, R.M.; Richard, F.; Sothilingam, V.; Garcia Garrido, M.; Le Bivic, A.; Seeliger, M.W.; et al. CRB2 acts as a modifying factor of CRB1-related retinal dystrophies in mice. Hum. Mol. Genet. 2014, 23, 3759–3771. [Google Scholar] [CrossRef] [Green Version]
- Quinn, P.M.; Mulder, A.A.; Henrique Alves, C.; Desrosiers, M.; de Vries, S.I.; Klooster, J.; Dalkara, D.; Koster, A.J.; Jost, C.R.; Wijnholds, J. Loss of CRB2 in Müller glial cells modifies a CRB1-associated retinitis pigmentosa phenotype into a Leber congenital amaurosis phenotype. Hum. Mol. Genet. 2019, 28, 105–123. [Google Scholar] [CrossRef] [Green Version]
- Kantardzhieva, A.; Gosens, I.; Alexeeva, S.; Punte, I.M.; Versteeg, I.; Krieger, E.; Neefjes-Mol, C.A.; Den Hollander, A.I.; Letteboer, S.J.F.F.; Klooster, J.; et al. MPP5 recruits MPP4 to the CRB1 complex in photoreceptors. Investig. Ophthalmol. Vis. Sci. 2005, 46, 2192–2201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Talib, M.; van Schooneveld, M.J.; van Genderen, M.M.; Wijnholds, J.; Florijn, R.J.; ten Brink, J.B.; Schalij-Delfos, N.E.; Dagnelie, G.; Cremers, F.P.M.; Wolterbeek, R.; et al. Genotypic and Phenotypic Characteristics of CRB1-Associated Retinal Dystrophies: A Long-Term Follow-up Study. Ophthalmology 2017, 124, 884–895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borrás, T. The pathway from genes to gene therapy in glaucoma: A review of possibilities for using genes as glaucoma drugs. Asia-Pacific J. Ophthalmol. 2017, 6, 80–93. [Google Scholar]
- Kong, D.H.; Kim, M.R.; Jang, J.H.; Na, H.J.; Lee, S. A review of anti-angiogenic targets for monoclonal antibody cancer therapy. Int. J. Mol. Sci. 2017, 18. [Google Scholar]
- Pechan, P.; Rubin, H.; Lukason, M.; Ardinger, J.; DuFresne, E.; Hauswirth, W.W.; Wadsworth, S.C.; Scaria, A. Novel anti-VEGF chimeric molecules delivered by AAV vectors for inhibition of retinal neovascularization. Gene Ther. 2009, 16, 10–16. [Google Scholar] [CrossRef]
- Heier, J.S.; Kherani, S.; Desai, S.; Dugel, P.; Kaushal, S.; Cheng, S.H.; Delacono, C.; Purvis, A.; Richards, S.; Le-Halpere, A.; et al. Intravitreous injection of AAV2-sFLT01 in patients with advanced neovascular age-related macular degeneration: A phase 1, open-label trial. Lancet 2017, 390, 50–61. [Google Scholar] [CrossRef]
- Parker, M.; Bellec, J.; McFarland, T.; Scripps, V.; Appukuttan, B.; Hartzell, M.; Yeager, A.; Hady, T.; Mitrophanous, K.A.; Stout, T.; et al. Suppression of neovascularization of donor corneas by transduction with equine infectious anemia virus-based lentiviral vectors expressing endostatin and angiostatin. Hum. Gene Ther. 2014, 25, 408–418. [Google Scholar] [CrossRef]
- Buchberger, A. The Therapeutic Utility of Factor I in the Treatment of Complement Dependent Pathophysiological Processes. Ph.D. Thesis, University of Leicester, Leicester, UK, 2016. [Google Scholar]
- Liu, Y.; Fortmann, S.D.; Shen, J.; Wielechowski, E.; Tretiakova, A.; Yoo, S.; Kozarsky, K.; Wang, J.; Wilson, J.M.; Campochiaro, P.A. AAV8-antiVEGFfab Ocular Gene Transfer for Neovascular Age-Related Macular Degeneration. Mol. Ther. 2018, 26, 542–549. [Google Scholar] [CrossRef] [Green Version]
- Cashman, S.M.; Ramo, K.; Kumar-Singh, R. A non membrane-targeted human soluble CD59 attenuates choroidal neovascularization in a model of age related macular degeneration. PLoS ONE 2011, 6. [Google Scholar] [CrossRef] [Green Version]
- Tomita, H.; Sugano, E.; Isago, H.; Hiroi, T.; Wang, Z.; Ohta, E.; Tamai, M. Channelrhodopsin-2 gene transduced into retinal ganglion cells restores functional vision in genetically blind rats. Exp. Eye Res. 2010, 90, 429–436. [Google Scholar] [CrossRef]
- Douar, A.M.; Bouquet, C.; Pruneau, D.; Chavas, J.; Dalkara, D.; Duebel, J.; Benosman, R.; Chenegros, G.; Picaud, S.; Sahel, J.; et al. 268. Optogenetic Engineering of Retinal Ganglion Cells with AAV2.7m8-ChrimsonR-tdTomato (GS030-DP) Is Well Tolerated and Induces Functional Responses to Light in Non-Human Primates. Mol. Ther. 2016, 24, S106–S107. [Google Scholar] [CrossRef] [Green Version]
- Lundstrom, K. Viral Vectors in Gene Therapy. Diseases 2018, 6, 42. [Google Scholar] [CrossRef] [Green Version]
- Trapani, I. Adeno-Associated Viral Vectors as a Tool for Large Gene Delivery to the Retina. Genes 2019, 10, 287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trapani, I.; Toriello, E.; de Simone, S.; Colella, P.; Iodice, C.; Polishchuk, E.V.; Sommella, A.; Colecchi, L.; Rossi, S.; Simonelli, F.; et al. Improved dual AAV vectors with reduced expression of truncated proteins are safe and effective in the retina of a mouse model of Stargardt disease. Hum. Mol. Genet. 2015, 24, 6811–6825. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McClements, M.E.; Barnard, A.R.; Singh, M.S.; Charbel Issa, P.; Jiang, Z.; Radu, R.A.; MacLaren, R.E. An AAV Dual Vector Strategy Ameliorates the Stargardt Phenotype in Adult Abca4-/- Mice. Hum. Gene Ther. 2019, 30, 590–600. [Google Scholar] [CrossRef]
- Kumar, N.; Stanford, W.; de Solis, C.; Aradhana; Abraham, N.D.; Dao, T.M.J.; Thaseen, S.; Sairavi, A.; Gonzalez, C.U.; Ploski, J.E. The development of an AAV-based CRISPR SaCas9 genome editing system that can be delivered to neurons in vivo and regulated via doxycycline and Cre-recombinase. Front. Mol. Neurosci. 2018, 11. [Google Scholar] [CrossRef] [Green Version]
- Hanlon, K.S.; Kleinstiver, B.P.; Garcia, S.P.; Zaborowski, M.P.; Volak, A.; Spirig, S.E.; Muller, A.; Sousa, A.A.; Tsai, S.Q.; Bengtsson, N.E.; et al. High levels of AAV vector integration into CRISPR-induced DNA breaks. Nat. Commun. 2019, 10. [Google Scholar] [CrossRef]
- Yu, W.; Mookherjee, S.; Chaitankar, V.; Hiriyanna, S.; Kim, J.W.; Brooks, M.; Ataeijannati, Y.; Sun, X.; Dong, L.; Li, T.; et al. Nrl knockdown by AAV-delivered CRISPR/Cas9 prevents retinal degeneration in mice. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef] [Green Version]
- Zhu, J.; Ming, C.; Fu, X.; Duan, Y.; Hoang, D.A.; Rutgard, J.; Zhang, R.; Wang, W.; Hou, R.; Zhang, D.; et al. Gene and mutation independent therapy via CRISPR-Cas9 mediated cellular reprogramming in rod photoreceptors. Cell Res. 2017, 27, 830–833. [Google Scholar] [CrossRef] [Green Version]
- Kim, E.; Koo, T.; Park, S.W.; Kim, D.; Kim, K.; Cho, H.Y.; Song, D.W.; Lee, K.J.; Jung, M.H.; Kim, S.; et al. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat. Commun. 2017, 8, 14500. [Google Scholar] [CrossRef] [Green Version]
- Jo, D.H.; Koo, T.; Cho, C.S.; Kim, J.H.; Kim, J.-S.; Kim, J.H. Long-Term Effects of In Vivo Genome Editing in the Mouse Retina Using Campylobacter jejuni Cas9 Expressed via Adeno-Associated Virus. Mol. Ther. 2019, 27, 130–136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perez-Pinera, P.; Kocak, D.D.; Vockley, C.M.; Adler, A.F.; Kabadi, A.M.; Polstein, L.R.; Thakore, P.I.; Glass, K.A.; Ousterout, D.G.; Leong, K.W.; et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat. Methods 2013, 10, 973–976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Konermann, S.; Brigham, M.D.; Trevino, A.E.; Joung, J.; Abudayyeh, O.O.; Barcena, C.; Hsu, P.D.; Habib, N.; Gootenberg, J.S.; Nishimasu, H.; et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 2015, 517, 583–588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lecomte, E.; Tournaire, B.; Cogné, B.; Dupont, J.B.; Lindenbaum, P.; Martin-Fontaine, M.; Broucque, F.; Robin, C.; Hebben, M.; Merten, O.W.; et al. Advanced characterization of DNA molecules in rAAV vector preparations by single-stranded virus next-generation sequencing. Mol. Ther.-Nucleic Acids 2015, 4, e260. [Google Scholar] [CrossRef]
- Bennicelli, J.; Wright, J.F.; Komaromy, A.; Jacobs, J.B.; Hauck, B.; Zelenaia, O.; Mingozzi, F.; Hui, D.; Chung, D.; Tonia, S.; et al. Reversal of Blindness in Animal Models of Leber Congenital Amaurosis Using Optimized AAV2-mediated Gene Transfer. Mol. Ther. 2010, 16, 458–465. [Google Scholar] [CrossRef]
- Mignon, C.; Sodoyer, R.; Werle, B. Antibiotic-free selection in biotherapeutics: Now and forever. Pathogens 2015, 4, 157–181. [Google Scholar] [CrossRef]
- Kay, M.A.; He, C.Y.; Chen, Z.Y. A robust system for production of minicircle DNA vectors. Nat. Biotechnol. 2010, 28, 1287–1289. [Google Scholar] [CrossRef] [Green Version]
- Schnödt, M.; Schmeer, M.; Kracher, B.; Krüsemann, C.; Espinosa, L.E.; Grünert, A.; Fuchsluger, T.; Rischmüller, A.; Schleef, M.; Büning, H. DNA Minicircle Technology Improves Purity of Adeno-associated Viral Vector Preparations. Mol. Ther.-Nucleic Acids 2016, 5, e355. [Google Scholar]
- Bishop, B.M.; Santin, A.D.; Quirk, J.G.; Hermonat, P.L. Role of terminal repeat GAGC trimer, the major Rep78 binding site, in adeno-associated virus DNA replication. Febs Lett. 1996, 397, 97–100. [Google Scholar] [CrossRef] [Green Version]
- Ryan, J.H.; Zolotukhin, S.; Muzyczka, N. Sequence requirements for binding of Rep68 to the adeno-associated virus terminal repeats. J. Virol. 1996, 70, 1542–1553. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.; Zeng, X.; Fan, Z.; Li, C.; McCown, T.; Samulski, R.J.; Xiao, X. Adeno-associated virus of a single-polarity DNA genome is capable of transduction in vivo. Mol. Ther. 2008, 16, 494–499. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.S.; Ponnazhagan, S.; Srivastava, A. Rescue and replication of adeno-associated virus type 2 as well as vector DNA sequences from recombinant plasmids containing deletions in the viral inverted terminal repeats: Selective encapsidation of viral genomes in progeny virions. J. Virol. 1996, 70, 1668–1677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.S.; Qing, K.; Ponnazhagan, S.; Srivastava, A. Adeno-associated virus type 2 DNA replication in vivo: Mutation analyses of the D sequence in viral inverted terminal repeats. J. Virol. 1997, 71, 3077–3082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwon, H.-J.; Qing, K.; Ponnazhagan, S.; Wang, X.-S.; Markusic, D.M.; Gupte, S.; Boye, S.; Srivastava, A. AAV D-sequence-mediated suppression of expression of a human major histocompatibility class II gene: Implications in the development of AAV vectors for modulating humoral immune response. Hum. Gene Ther. 2020, 31, 565–574. [Google Scholar] [CrossRef] [PubMed]
- Samulski, R.J.; Srivastava, A.; Berns, K.I.; Muzyczka, N. Rescue of adeno-associated virus from recombinant plasmids: Gene correction within the terminal repeats of AAV. Cell 1983, 33, 135–143. [Google Scholar] [CrossRef]
- Zhou, Q.; Tian, W.; Liu, C.; Lian, Z.; Dong, X.; Wu, X. Deletion of the B-B’ and C-C’ regions of inverted terminal repeats reduces rAAV productivity but increases transgene expression. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef]
- McAlister, V.J.; Owens, R.A. Substitution of adeno-associated virus Rep protein binding and nicking sites with human chromosome 19 sequences. Virol. J. 2010, 7, 218. [Google Scholar] [CrossRef] [Green Version]
- Crémisi, C.; Pignatti, P.F.; Yaniv, M. Random location and absence of movement of the nucleosomes on SV 40 nucleoprotein complex isolated from infected cells. Biochem. Biophys. Res. Commun. 1976, 73, 548–554. [Google Scholar] [CrossRef]
- Okada, T.; Uchibori, R.; Iwata-Okada, M.; Takahashi, M.; Nomoto, T.; Nonaka-Sarukawa, M.; Ito, T.; Liu, Y.; Mizukami, H.; Kume, A.; et al. A histone deacetylase inhibitor enhances recombinant adeno-associated virus-mediated gene expression in tumor cells. Mol. Ther. 2006, 13, 738–746. [Google Scholar] [CrossRef]
- Crémisi, C.; Chestier, A.; Yaniv, M. Preferential association of newly synthesized histones with replicating SV40 DNA. Cell 1977, 12, 947–951. [Google Scholar] [CrossRef]
- Mano, M.; Ippodrino, R.; Zentilin, L.; Zacchigna, S.; Giacca, M. Genome-wide RNAi screening identifies host restriction factors critical for in vivo AAV transduction. Proc. Natl. Acad. Sci. USA 2015, 112, 11276–11281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Provost, N.; Le Meur, G.; Weber, M.; Mendes-Madeira, A.; Podevin, G.; Cherel, Y.; Colle, M.A.; Deschamps, J.Y.; Moullier, P.; Rolling, F. Biodistribution of rAAV vectors following intraocular administration: Evidence for the presence and persistence of vector DNA in the optic nerve and in the brain. Mol. Ther. 2005, 11, 275–283. [Google Scholar] [CrossRef] [PubMed]
- Miller, D.G.; Petek, L.M.; Russell, D.W. Adeno-associated virus vectors integrate at chromosome breakage sites. Nat. Genet. 2004, 36, 767–773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakai, H.; Yant, S.R.; Storm, T.A.; Fuess, S.; Meuse, L.; Kay, M.A. Extrachromosomal recombinant adeno-associated virus vector genomes are primarily responsible for stable liver transduction in vivo. J. Virol. 2001, 75, 6969–6976. [Google Scholar] [CrossRef] [Green Version]
- Colella, P.; Ronzitti, G.; Mingozzi, F. Emerging Issues in AAV-Mediated In Vivo Gene Therapy. Mol. Ther.-Methods Clin. Dev. 2018, 8, 87–104. [Google Scholar] [CrossRef] [Green Version]
- Nakai, H.; Montini, E.; Fuess, S.; Storm, T.A.; Grompe, M.; Kay, M.A. AAV serotype 2 vectors preferentially integrate into active genes in mice. Nat. Genet. 2003, 34, 297–302. [Google Scholar] [CrossRef]
- Ceiler, J.; Afzal, S.; Leuchs, B.; Fronza, R.; Lulay, C.; Büning, H.; Schmidt, M.; Gil-Farina, I. Wild-Type and Recombinant AAV Integration in Human Cardiomyocytes: Focus on Mitochondrial Genome. In Proceedings of the AAV Vector Biology, 10 May 2017; Available online: https://www.researchgate.net/publication/316699961_Wild-Type_and_Recombinant_AAV_Integration_in_Human_Cardiomyocytes_Focus_on_Mitochondrial_Genome (accessed on 10 June 2020).
- Gil-Farina, I.; Fronza, R.; Kaeppel, C.; Lopez-Franco, E.; Ferreira, V.; D’Avola, D.; Benito, A.; Prieto, J.; Petry, H.; Gonzalez-Aseguinolaza, G.; et al. Recombinant AAV Integration Is Not Associated with Hepatic Genotoxicity in Nonhuman Primates and Patients. Mol. Ther. 2016, 24, 1100–1105. [Google Scholar] [CrossRef] [Green Version]
- Maeder, M.L.; Stefanidakis, M.; Wilson, C.J.; Baral, R.; Barrera, L.A.; Bounoutas, G.S.; Bumcrot, D.; Chao, H.; Ciulla, D.M.; DaSilva, J.A.; et al. Development of a gene-editing approach to restore vision loss in Leber congenital amaurosis type 10. Nat. Med. 2019, 25, 229–233. [Google Scholar] [CrossRef]
- Wang, Z.; Lisowski, L.; Finegold, M.J.; Nakai, H.; Kay, M.A.; Grompe, M. AAV vectors containing rDNA homology display increased chromosomal integration and transgene persistence. Mol. Ther. 2012, 20, 1902–1911. [Google Scholar] [CrossRef] [Green Version]
- Hirsch, M.L. Adeno-associated virus inverted terminal repeats stimulate gene editing. Gene Ther. 2015, 22, 190–195. [Google Scholar] [CrossRef] [Green Version]
- Ronzitti, G.; Bortolussi, G.; van Dijk, R.; Collaud, F.; Charles, S.; Leborgne, C.; Vidal, P.; Martin, S.; Gjata, B.; Sola, M.S.; et al. A translationally optimized AAV-UGT1A1 vector drives safe and long-lasting correction of Crigler-Najjar syndrome. Mol. Ther.-Methods Clin. Dev. 2016, 3, 16049. [Google Scholar] [CrossRef]
- McCarty, D.M.; Monahan, P.E.; Samulski, R.J. Self-complementary recombinant adeno-associated virus (scAAV) vectors promote efficient transduction independently of DNA synthesis. Gene Ther. 2001, 8, 1248–1254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCarty, D.M. Self-complementary AAV vectors; advances and applications. Mol. Ther. 2008, 16, 1648–1656. [Google Scholar] [CrossRef] [PubMed]
- Koilkonda, R.D.; Chou, T.H.; Porciatti, V.; Hauswirth, W.W.; Guy, J. Induction of rapid and highly efficient expression of the human ND4 complex I subunit in the mouse visual system by self-complementary adeno-associated virus. Arch. Ophthalmol. 2010, 128, 876–883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yokoi, K.; Kachi, S.; Zhang, H.S.; Gregory, P.D.; Spratt, S.K.; Samulski, R.J.; Campochiaro, P.A. Ocular gene transfer with self-complementary AAV vectors. Investig. Ophthalmol. Vis. Sci. 2007, 48, 3324–3328. [Google Scholar] [CrossRef] [Green Version]
- Wright, J.F.; Zelenaia, O. Vector characterization methods for quality control testing of recombinant adeno-associated viruses. Methods Mol. Biol. 2011, 737, 247–278. [Google Scholar]
- François, A.; Bouzelha, M.; Lecomte, E.; Broucque, F.; Penaud-Budloo, M.; Adjali, O.; Moullier, P.; Blouin, V.; Ayuso, E. Accurate Titration of Infectious AAV Particles Requires Measurement of Biologically Active Vector Genomes and Suitable Controls. Mol. Ther.-Methods Clin. Dev. 2018, 10, 223–236. [Google Scholar]
- Couto, L.; Buchlis, G.; Farjo, R.; High, K.A. Poster C0048 ARVO: Potency Assay for AAV Vector Encoding Retinal Pigment Epithelial 65 Protein. Investig. Ophthalmol. Vis. Sci. 2016, 57, 1. [Google Scholar]
- Patrício, M.I.; Barnard, A.R.; Cox, C.I.; Blue, C.; MacLaren, R.E. The Biological Activity of AAV Vectors for Choroideremia Gene Therapy Can Be Measured by In Vitro Prenylation of RAB6A. Mol. Ther.-Methods Clin. Dev. 2018, 9, 288–295. [Google Scholar]
- Wang, L.; Xiao, R.; Andres-Mateos, E.; Vandenberghe, L.H. Single stranded adeno-associated virus achieves efficient gene transfer to anterior segment in the mouse eye. PLoS ONE 2017, 12. [Google Scholar] [CrossRef]
- Borrás, T.; Xue, W.; Choi, V.W.; Bartlett, J.S.; Li, G.; Samulski, R.J.; Chisolm, S.S. Mechanisms of AAV transduction in glaucoma-associated human trabecular meshwork cells. J. Gene Med. 2006, 8, 589–602. [Google Scholar] [CrossRef] [PubMed]
- Ellis, B.L.; Hirsch, M.L.; Barker, J.C.; Connelly, J.P.; Steininger, R.J.; Porteus, M.H. A survey of ex vivo/in vitro transduction efficiency of mammalian primary cells and cell lines with Nine natural adeno-associated virus (AAV1-9) and one engineered adeno-associated virus serotype. Virol. J. 2013, 10, 74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonzalez-Cordero, A.; Goh, D.; Kruczek, K.; Naeem, A.; Fernando, M.; Kleine Holthaus, S.M.; Takaaki, M.; Blackford, S.J.I.; Kloc, M.; Agundez, L.; et al. Assessment of AAV Vector Tropisms for Mouse and Human Pluripotent Stem Cell-Derived RPE and Photoreceptor Cells. Hum. Gene Ther. 2018, 29, 1124–1139. [Google Scholar] [CrossRef] [PubMed]
- Sayyad, Z.; Sirohi, K.; Radha, V.; Swarup, G. 661W is a retinal ganglion precursor-like cell line in which glaucoma-associated optineurin mutants induce cell death selectively. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [Green Version]
- Tan, E.; Ding, X.-Q.; Saadi, A.; Agarwal, N.; Naash, M.I.; Al-Ubaidi, M.R. Expression of cone-photoreceptor-specific antigens in a cell line derived from retinal tumors in transgenic mice. Investig. Ophthalmol. Vis. Sci. 2004, 45, 764–768. [Google Scholar] [CrossRef] [Green Version]
- Ryals, R.C.; Boye, S.L.; Dinculescu, A.; Hauswirth, W.W.; Boye, S.E. Quantifying transduction efficiencies of unmodified and tyrosine capsid mutant AAV vectors in vitro using two ocular cell lines. Mol. Vis. 2011, 17, 1090–1102. [Google Scholar]
- McDougald, D.S.; Duong, T.T.; Palozola, K.C.; Marsh, A.; Papp, T.E.; Mills, J.A.; Zhou, S.; Bennett, J. CRISPR Activation Enhances In Vitro Potency of AAV Vectors Driven by Tissue-Specific Promoters. Mol. Ther.-Methods Clin. Dev. 2019, 13, 380–389. [Google Scholar] [CrossRef] [Green Version]
- Drivas, T.G.; Bennett, J. Compositions and Methods for Treatment of Disorders Related to CEP290. U.S. Patent 10301366B2, 22 January 2015. [Google Scholar]
- Katoh, Y.; Michisaka, S.; Nozaki, S.; Funabashi, T.; Hirano, T.; Takei, R.; Nakayama, K. Practical method for targeted disruption of ciliarelated genes by using CRISPR/Cas9-mediated, homology-independent knock-in system. Mol. Biol. Cell 2017, 28, 898–906. [Google Scholar] [CrossRef]
- Klimczak, R.R.; Koerber, J.T.; Dalkara, D.; Flannery, J.G.; Schaffer, D.V. A novel adeno-associated viral variant for efficient and selective intravitreal transduction of rat Müller cells. PLoS ONE 2009, 4, e7467. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Ma, Z.; Zou, W.; Guo, H.; Liu, M.; Ma, Y.; Zhang, L. The Appropriate Marker for Astrocytes: Comparing the Distribution and Expression of Three Astrocytic Markers in Different Mouse Cerebral Regions. Biomed Res. Int. 2019, 2019, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Vecino, E.; Rodriguez, F.D.; Ruzafa, N.; Pereiro, X.; Sharma, S.C. Glia-neuron interactions in the mammalian retina. Prog. Retin. Eye Res. 2016, 51, 1–40. [Google Scholar] [CrossRef] [Green Version]
- Lawrence, J.M.; Singhal, S.; Bhatia, B.; Keegan, D.J.; Reh, T.A.; Luthert, P.J.; Khaw, P.T.; Limb, G.A. MIO-M1 Cells and Similar Müller Glial Cell Lines Derived from Adult Human Retina Exhibit Neural Stem Cell Characteristics. Stem Cells 2007, 25, 2033–2043. [Google Scholar] [CrossRef] [PubMed]
- Limb, G.A.; Salt, T.E.; Munro, P.M.G.; Moss, S.E.; Khaw, P.T. In vitro characterization of a spontaneously immortalized human Müller cell line (MIO-M1). Investig. Ophthalmol. Vis. Sci. 2002, 43, 864–869. [Google Scholar] [PubMed]
- Koch, P.; de Greef, S.; Chtarto, A.; Kemp, T.; Bockstae, O.; Velu, T.; Caspers, L.; Tenenbaum, L. ARVO abstract: In vitro comparison of AAV–mediated eGFP gene transfer on Müller cells and ARPE cells. Investig. Ophthalmol. Vis. Sci. 2004, 45. [Google Scholar]
- Mellough, C.B.; Sernagor, E.; Moreno-Gimeno, I.; Steel, D.H.W.; Lako, M. Efficient stage-specific differentiation of human pluripotent stem cells toward retinal photoreceptor cells. Stem Cells 2012, 30, 673–686. [Google Scholar] [CrossRef]
- Cereso, N.; Pequignot, M.O.; Robert, L.; Becker, F.; De Luca, V.; Nabholz, N.; Rigau, V.; De Vos, J.; Hamel, C.P.; Kalatzis, V. Proof of concept for AAV2/5-mediated gene therapy in iPSC-derived retinal pigment epithelium of a choroideremia patient. Mol. Ther.-Methods Clin. Dev. 2014, 1, 14011. [Google Scholar] [CrossRef]
- Duong, T.T.; Vasireddy, V.; Ramachandran, P.; Herrera, P.S.; Leo, L.; Merkel, C.; Bennett, J.; Mills, J.A. Use of induced pluripotent stem cell models to probe the pathogenesis of Choroideremia and to develop a potential treatment. Stem Cell Res. 2018, 27, 140–150. [Google Scholar] [CrossRef]
- Brydon, E.M.; Bronstein, R.; Buskin, A.; Lako, M.; Pierce, E.A.; Fernandez-Godino, R. AAV-mediated gene augmentation therapy restores critical functions in mutant iPSC-derived PRPF31+/- cells. bioRxiv 2019, 729160. [Google Scholar] [CrossRef]
- Wright, J.F.; Sumaroka, M. Rab Escort Protein Potency Assay. U.S. Patent 201662418637P, 11 September 2019. [Google Scholar]
- Garita-Hernandez, M.; Guibbal, L.; Toualbi, L.; Routet, F.; Chaffiol, A.; Winckler, C.; Harinquet, M.; Robert, C.; Fouquet, S.; Bellow, S.; et al. Optogenetic light sensors in human retinal organoids. Front. Neurosci. 2018, 12. [Google Scholar] [CrossRef] [PubMed]
- Garita-Hernandez, M.; Routet, F.; Guibbal, L.; Khabou, H.; Toualbi, L.; Riancho, L.; Reichman, S.; Duebel, J.; Sahel, J.A.; Goureau, O.; et al. AAV-mediated gene delivery to 3D retinal organoids derived from human induced pluripotent stem cells. Int. J. Mol. Sci. 2020, 21, 994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dudek, A.M.; Pillay, S.; Puschnik, A.S.; Nagamine, C.M.; Cheng, F.; Qiu, J.; Carette, J.E.; Vandenberghe, L.H. An Alternate Route for Adeno-associated Virus (AAV) Entry Independent of AAV Receptor. J. Virol. 2018, 92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akl, M.R.; Nagpal, P.; Ayoub, N.M.; Tai, B.; Prabhu, S.A.; Capac, C.M.; Gliksman, M.; Goy, A.; Suh, K.S. Molecular and clinical significance of fibroblast growth factor 2 (FGF2/bFGF) in malignancies of solid and hematological cancers for personalized therapies. Oncotarget 2016, 7, 44735–44762. [Google Scholar] [CrossRef] [Green Version]
- ThermoScientific Growth Factors in HyClone Cell Culture Serum. Available online: https://static.thermoscientific.com/images/D22225~.pdf (accessed on 11 June 2020).
- Qing, K.; Mah, C.; Hansen, J.; Zhou, S.; Dwarki, V.; Srivastava, A. Human fibroblast growth factor receptor 1 is a co-receptor for infection by adeno-associated virus 2. Nat. Med. 1999, 5, 71–77. [Google Scholar] [CrossRef]
- Capowski, E.E.; Samimi, K.; Mayerl, S.J.; Phillips, M.J.; Pinilla, I.; Howden, S.E.; Saha, J.; Jansen, A.D.; Edwards, K.L.; Jager, L.D.; et al. Reproducibility and staging of 3D human retinal organoids across multiple pluripotent stem cell lines. Development 2019, 146. [Google Scholar] [CrossRef] [Green Version]
- Mellough, C.B.; Collin, J.; Queen, R.; Hilgen, G.; Dorgau, B.; Zerti, D.; Felemban, M.; White, K.; Sernagor, E.; Lako, M. Systematic Comparison of Retinal Organoid Differentiation from Human Pluripotent Stem Cells Reveals Stage Specific, Cell Line, and Methodological Differences. Stem Cells Transl. Med. 2019, 8, 694–706. [Google Scholar] [CrossRef] [Green Version]
- Achberger, K.; Haderspeck, J.C.; Kleger, A.; Liebau, S. Stem cell-based retina models. Adv. Drug Deliv. Rev. 2019, 140, 33–50. [Google Scholar] [CrossRef]
- Wiley, L.A.; Burnight, E.R.; Kaalberg, E.E.; Jiao, C.; Riker, M.J.; Halder, J.A.; Luse, M.A.; Han, I.C.; Russell, S.R.; Sohn, E.H.; et al. Assessment of Adeno-Associated Virus Serotype Tropism in Human Retinal Explants. Hum. Gene Ther. 2018, 29, 424–436. [Google Scholar] [CrossRef]
- Khabou, H.; Garita-Hernandez, M.; Chaffiol, A.; Reichman, S.; Jaillard, C.; Brazhnikova, E.; Bertin, S.; Forster, V.; Desrosiers, M.; Winckler, C.; et al. Noninvasive gene delivery to foveal cones for vision restoration. Jci Insight 2018, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hickey, D.G.; Edwards, T.L.; Barnard, A.R.; Singh, M.S.; de Silva, S.R.; McClements, M.E.; Flannery, J.G.; Hankins, M.W.; MacLaren, R.E. Tropism of engineered and evolved recombinant AAV serotypes in the rd1 mouse and ex vivo primate retina. Gene Ther. 2017, 24, 787–800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buck, T.M.; Pellissier, L.P.; Vos, R.M.; van Dijk, E.H.C.C.; Boon, C.J.F.; Wijnholds, J. AAV serotype testing on cultured human donor retinal explants. In Methods in Molecular Biology; Boon, C.J.F., Wijnholds, J., Eds.; Humana Press: New York, NY, USA, 2018; Volume 1715, pp. 275–288. ISBN 978-1-4939-7522-8. [Google Scholar]
- Reid, C.A.; Ertel, K.J.; Lipinski, D.M. Improvement of photoreceptor targeting via intravitreal delivery in mouse and human retina using combinatory rAAV2 capsid mutant vectors. Investig. Ophthalmol. Vis. Sci. 2017, 58, 6429–6439. [Google Scholar] [CrossRef] [PubMed]
- Buck, T.M.; Quinn, P.M.; Celso Alves, H.; Van Dijk, E.; Ohonin, C.; Boon, C.J.F.; Wijnholds, J. Potency assay for AAV-gene vectors in human iPSCs-derived retinas and donor retinas | IOVS | ARVO Journals. Investig. Ophthalmol. Vis. Sci. 2017, 58, 4093. [Google Scholar]
- Singh, H.P.; Wang, S.; Stachelek, K.; Lee, S.; Reid, M.W.; Thornton, M.E.; Craft, C.M.; Grubbs, B.H.; Cobrinik, D. Developmental stage-specific proliferation and retinoblastoma genesis in RB-deficient human but not mouse cone precursors. Proc. Natl. Acad. Sci. USA 2018, 115, E9391–E9400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petit, L.; Ma, S.; Cheng, S.-Y.; Gao, G.; Punzo, C. Rod Outer Segment Development Influences AAV-Mediated Photoreceptor Transduction After Subretinal Injection. Hum. Gene Ther. 2017, 28, 464–481. [Google Scholar] [CrossRef]
- Surace, E.M.; Auricchio, A.; Reich, S.J.; Glover, E.; Pineles, S.; Tang, W.; O’Connor, E.; Lyubarsky, A.; Savchenko, A.; Pugh, E.N.; et al. Delivery of Adeno-Associated Virus Vectors to the Fetal Retina: Impact of Viral Capsid Proteins on Retinal Neuronal Progenitor Transduction. J. Virol. 2003, 77, 7957–7963. [Google Scholar] [CrossRef] [Green Version]
- Xiong, W.; Cepko, C. Distinct Expression Patterns of AAV8 Vectors with Broadly Active Promoters from Subretinal Injections of Neonatal Mouse Eyes at Two Different Ages. Adv. Exp. Med. Biol. 2016, 854, 501–507. [Google Scholar]
- Matsumoto, B.; Blanks, J.C.; Ryan, S.J. Topographic variations in the rabbit and primate internal limiting membrane. | IOVS | ARVO Journals. Investig. Ophthalmol. Vis. Sci. 1984, 2, 71–82. [Google Scholar]
- Timmers, A.M.; Newmark, J.A.; Turunen, H.T.; Farivar, T.; Liu, J.; Song, C.; Ye, G.J.; Pennock, S.; Gaskin, C.; Knop, D.R.; et al. Ocular Inflammatory Response to Intravitreal Injection of Adeno-Associated Virus Vector: Relative Contribution of Genome and Capsid. Hum. Gene Ther. 2020, 31, 80–89. [Google Scholar] [CrossRef]
- Dalkara, D.; Kolstad, K.D.; Caporale, N.; Visel, M.; Klimczak, R.R.; Schaffer, D.V.; Flannery, J.G. Inner limiting membrane barriers to aav-mediated retinal transduction from the vitreous. Mol. Ther. 2009, 17, 2096–2102. [Google Scholar] [CrossRef] [Green Version]
- Cehajic-Kapetanovic, J.; Milosavljevic, N.; Bedford, R.A.; Lucas, R.J.; Bishop, P.N. Efficacy and Safety of Glycosidic Enzymes for Improved Gene Delivery to the Retina following Intravitreal Injection in Mice. Mol. Ther.-Methods Clin. Dev. 2018, 9, 192–202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, H.; Bush, R.A.; Zeng, Y.; Qian, H.; Wu, Z.; Sieving, P.A. Trans-ocular Electric Current In Vivo Enhances AAV-Mediated Retinal Gene Transduction after Intravitreal Vector Administration. Mol. Ther.-Methods Clin. Dev. 2019, 13, 77–85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dias, M.S.; Araujo, V.G.; Vasconcelos, T.; Li, Q.; Hauswirth, W.W.; Linden, R.; Petrs-Silva, H. Retina transduction by rAAV2 after intravitreal injection: Comparison between mouse and rat. Gene Ther. 2019, 26, 479–490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolstad, K.D.; Dalkara, D.; Guerin, K.; Visel, M.; Hoffmann, N.; Schaffer, D.V.; Flannery, J.G. Changes in adeno-associated virus-mediated gene delivery in retinal degeneration. Hum. Gene Ther. 2010, 21, 571–578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Charbel Issa, P.; de Silva, S.R.; Lipinski, D.M.; Singh, M.S.; Mouravlev, A.; You, Q.; Barnard, A.R.; Hankins, M.W.; During, M.J.; MacLaren, R.E. Assessment of Tropism and Effectiveness of New Primate-Derived Hybrid Recombinant AAV Serotypes in the Mouse and Primate Retina. PLoS ONE 2013, 8, e0060361. [Google Scholar] [CrossRef] [Green Version]
- Zeng, Y.; Qian, H.; Wu, Z.; Marangoni, D.; Sieving, P.A.; Bush, R.A. AAVrh-10 transduces outer retinal cells in rodents and rabbits following intravitreal administration. Gene Ther. 2019, 26, 386–398. [Google Scholar] [CrossRef] [Green Version]
- Giove, T.J.; Sena-Esteves, M.; Eldred, W.D. Transduction of the inner mouse retina using AAVrh8 and AAVrh10 via intravitreal injection. Exp. Eye Res. 2010, 91, 652–659. [Google Scholar] [CrossRef] [Green Version]
- Lovric, J.; Mano, M.; Zentilin, L.; Eulalio, A.; Zacchigna, S.; Giacca, M. Terminal differentiation of cardiac and skeletal myocytes induces permissivity to AAV transduction by relieving inhibition imposed by DNA damage response proteins. Mol. Ther. 2012, 20, 2087–2097. [Google Scholar] [CrossRef] [Green Version]
- Moreno, M.L.; Mérida, S.; Bosch-Morell, F.; Miranda, M.; Villar, V.M. Autophagy dysfunction and oxidative stress, two related mechanisms implicated in retinitis pigmentosa. Front. Physiol. 2018, 9, 1008. [Google Scholar] [CrossRef]
- Blasiak, J.; Piechota, M.; Pawlowska, E.; Szatkowska, M.; Sikora, E.; Kaarniranta, K. Cellular senescence in age-related macular degeneration: Can autophagy and DNA damage response play a role? Oxid. Med. Cell. Longev. 2017, 2017. [Google Scholar] [CrossRef]
- D’Adda Di Fagagna, F. Living on a break: Cellular senescence as a DNA-damage response. Nat. Rev. Cancer 2008, 8, 512–522. [Google Scholar] [CrossRef]
- Lotery, A.J.; Yang, G.S.; Mullins, R.F.; Russell, S.R.; Schmidt, M.; Stone, E.M.; Lindbloom, J.D.; Chiorini, J.A.; Kotin, R.M.; Davidson, B.L. Adeno-Associated Virus Type 5: Transduction Efficiency and Cell-Type Specificity in the Primate Retina. Hum. Gene Ther. 2003, 14, 1663–1671. [Google Scholar] [CrossRef] [PubMed]
- Gao, G.-P.; Auricchio, A.; Wang, L.; Johnston, J.; Hammill, D.; Raper, S.E.; Grant, R.; Bennett, J.; Wilson, J.M.; Gao, G.-P.; et al. 561. Evaluation of AAV Vectors Based on Serotypes 1,2 and 5 in Non-Human Primate Muscle, Liver and Retina. Mol. Ther. 2002, 5, S184. [Google Scholar]
- Vandenberghe, L.H.; Bell, P.; Maguire, A.M.; Xiao, R.; Hopkins, T.B.; Grant, R.; Bennett, J.; Wilson, J.M. AAV9 targets cone photoreceptors in the nonhuman primate retina. PLoS ONE 2013, 8, e53463. [Google Scholar] [CrossRef]
- Ramachandran, P.S.; Lee, V.; Wei, Z.; Song, J.Y.; Casal, G.; Cronin, T.; Willett, K.; Huckfeldt, R.; Morgan, J.I.W.; Aleman, T.S.; et al. Evaluation of Dose and Safety of AAV7m8 and AAV8BP2 in the Non-Human Primate Retina. Hum. Gene Ther. 2017, 28, 154–167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carvalho, L.S.; Xiao, R.; Wassmer, S.J.; Langsdorf, A.; Zinn, E.; Pacouret, S.; Shah, S.; Comander, J.I.; Kim, L.A.; Lim, L.; et al. Synthetic Adeno-Associated Viral Vector Efficiently Targets Mouse and Nonhuman Primate Retina in Vivo. Hum. Gene Ther. 2018, 29, 771–784. [Google Scholar] [CrossRef]
- Boye, S.E.; Alexander, J.J.; Boye, S.L.; Witherspoon, C.D.; Sandefer, K.J.; Conlon, T.J.; Erger, K.; Sun, J.; Ryals, R.; Chiodo, V.A.; et al. The Human Rhodopsin Kinase Promoter in an AAV5 Vector Confers Rod- and Cone-Specific Expression in the Primate Retina. Hum. Gene Ther. 2012, 23, 1101–1115. [Google Scholar] [CrossRef] [Green Version]
- Yin, L.; Greenberg, K.; Hunter, J.J.; Dalkara, D.; Kolstad, K.D.; Masella, B.D.; Wolfe, R.; Visel, M.; Stone, D.; Libby, R.T.; et al. Intravitreal injection of AAV2 transduces macaque inner retina. Investig. Ophthalmol. Vis. Sci. 2011, 52, 2775–2783. [Google Scholar] [CrossRef]
- Takahashi, K.; Igarashi, T.; Miyake, K.; Kobayashi, M.; Yaguchi, C.; Iijima, O.; Yamazaki, Y.; Katakai, Y.; Miyake, N.; Kameya, S.; et al. Improved Intravitreal AAV-Mediated Inner Retinal Gene Transduction after Surgical Internal Limiting Membrane Peeling in Cynomolgus Monkeys. Mol. Ther. 2017, 25, 296–302. [Google Scholar] [CrossRef] [Green Version]
- Xiong, W.; Wu, D.M.; Xue, Y.; Wang, S.K.; Chung, M.J.; Ji, X.; Rana, P.; Zhao, S.R.; Mai, S.; Cepko, C.L. AAV cis-regulatory sequences are correlated with ocular toxicity. Proc. Natl. Acad. Sci. USA 2019, 116, 5785–5794. [Google Scholar] [CrossRef] [Green Version]
- Peirson, S.N.; Brown, L.A.; Pothecary, C.A.; Benson, L.A.; Fisk, A.S. Light and the laboratory mouse. J. Neurosci. Methods 2018, 300, 26–36. [Google Scholar] [CrossRef] [PubMed]
- Khabou, H.; Cordeau, C.; Pacot, L.; Fisson, S.; Dalkara, D. Dosage Thresholds and Influence of Transgene Cassette in Adeno-Associated Virus-Related Toxicity. Hum. Gene Ther. 2018, 29, 1235–1241. [Google Scholar] [CrossRef] [PubMed]
- Mingozzi, F.; High, K.A. Therapeutic in vivo gene transfer for genetic disease using AAV: Progress and challenges. Nat. Rev. Genet. 2011, 12, 341–355. [Google Scholar] [CrossRef] [PubMed]






| Disease | Year | Product | Capsid | Promoter | Intron | Other | Gene | PolyA | Production |
|---|---|---|---|---|---|---|---|---|---|
| LCA | 2007 | hRPE65v2 | AAV2 | CAG | β-globin | RPE65 | bGH | HEK293 | |
| LCA | 2007 | AAV-RPE65 | AAV2 | CB-SB | RPE65 | SV40 | HEK293 | ||
| LCA | 2008 | tgAAG76 | AAV2 | hRPE65 | RPE65 | bGH | B50, helper adenovirus | ||
| LCA | 2011 | HORA-RPE65 | AAV4 | hRPE65 | RPE65 | bGH | HEK293 | ||
| LCA | 2016 | OPTIRPE | AAV5 | NA65 | SV40 | RPE65 | SV40 | HEK293 | |
| AMD | 2009 | sFLT01 | AAV2 | CAG | β-globin | sFLT01 | bGH | HEK293 | |
| AMD | 2011 | OXB-201 | EIAV | CMV | IRES + WPRE | Endo+ Angio | SIN-LTR | ? | |
| AMD | 2011 | AAV.sFlt-1 | AAV2 | CMV | Chimeric intron | sFlt-1 | SV40 | HEK293 | |
| AMD | 2017 | RGX-314 | AAV8 | CAG/CB7 | β-globin | aVEGFAfabH.F2A .aVEGFfabL | rabbit β-globin | ? | |
| AMD | 2018 | HMR59 | AAV2 | CAG | β-globinSD/SA | sCD59 | bGH | ? | |
| AMD | 2018 | ADVM-022 | AAV2-7m8 | CMV | β-globinSD IgSA | TLP-eMLP | sFLT01co | Sf9 | |
| AMD | 2019 | GT005 | AAV2 | CBA | β-globin | WPRE | CFI | bGH | ? |
| LHON | 2010 | AAV2-ND4 | AAV2 | CMV | 5′UTR COX10 | 3′UTR COX10(MTS) | ND4 | bGH | HEK293, HSV1-rc/ΔUL2 |
| LHON | 2011 | scAAV2-P1ND4v2 | AAV2-tYF | smCBA | ATP1(MTS); WPRE | ND4 | bGH | HEK293 | |
| LHON | 2014 | GSO10 | AAV2 | CMV | β-globin | COX10(MTS) | ND4 | 3′COX10 | HEK293 |
| Stargardt | 2011 | SAR422459 | EIAV | CMV | ABCA4 | SIN-LTR | HEK293 | ||
| CHM | 2011 | AAV2.REP1 | AAV2 | CAG | β-globinSD/SA | WPRE | CHM | bGH | HEK293 |
| CHM | 2015 | AAV2.REP1 | AAV2 | CAG | β-globin | CHM | bGH | HEK293 | |
| RP | 2011 | AAV2.MERTK | AAV2 | hVMD2 | SV40SD/SA | MERTK | SV40. bGH | HEK293 | |
| Usher | 2012 | UshStat | EIAV | CMV | WPRE | MYO7A | SIN-LTR | HEK293 | |
| Usher | 2018 | QR-421a | AON-USH2A | Synthetic | |||||
| LCA | 2019 | EDIT-101 | AAV5 | U6; hGRK1 | SV40SD/SA | gRNA-CEP290 | SaCas9 | Synthetic | HEK293 |
| LCA | 2019 | AAV5.GUCY2D | AAV5 | hGRK1 | SV40SD/SA | GUCY2D | bGH | HeLaS3 | |
| XLR | 2015 | AAV2-tYF.RS1 | AAV2-tYF | smCB | β-globinSD/SA | WPRE | RS1 | SV40 | rHSV/sBHK |
| XLR | 2017 | scAAV8-RS1 | AAV8 | hRS1 | RS1 | IRBP enhancer | RS1 | Human β-globin | HEK293 |
| ACHM | 2015 | AAV2-tYF.CNGB3 | AAV2-tYF | PR1.7 | SV40SD/SA | CNGB3 | SV40 | rHSV/sBHK | |
| ACHM | 2015 | AAV.CNGA3 | AAV8 | hCAR | WPREm | CNGA3 | bGH | HEK293 | |
| ACHM | 2016 | AAV8.CNGA3 | AAV8 | hG1.7 | CNGA3 | SV40 | HEK293 | ||
| ACHM | 2016 | AAV8.CNGB3 | AAV8 | hCAR | CNGB3 | SV40 | HEK293 | ||
| ACHM | 2019 | AGTC-402 | AAV2-tYF | PR1.7 | SV40SD/SA | CNGA3 | SV40 | rHSV/sBHK | |
| RP | 2017 | AAV8.RPGR | AAV8 | hGRK1 | RPGRco-ORF15 | bGH | HEK293 | ||
| RP | 2017 | AAV-RPGR | AAV5 | hGRK1 | SV40SD/SA | RPGRco-ORF15-Long | SV40 | HEK293 | |
| RP | 2017 | AGTC-501 | AAV2-tYF | hGRK1 | SV40SD/SA | RPGRco-ORF15 | SV40 | rHSV/sBHK | |
| RP | 2017 | RST-001 | AAV2 | CAG | β-globinSD/SA | WPRE | Chop2/ChR2 | bGH | HEK293 |
| RP | 2017 | GS030 | AAV2-7m8 | CAG | ChrimsonR-tdT | bGH | ? | ||
| RP | 2020 | BSO1 | AAV? | ? | Chr90-FP | ? | ? | ||
| RP | 2017 | AAV5.PDE6B | AAV5 | hGRK1 | PED6B | bGH | HEK293 | ||
| RP | 2017 | CPK850 | scAAV8 | sRLBP1 | mSV40SD/SA | RLBP1 | SV40 | HEK293 |
| Ubiquitous Promoters | Size (bp) | Origin, Cell Expression, Strength | References |
|---|---|---|---|
| CAGGS aka CBA or CAG | 1600 | Ubiquitous, +++. Cytomegalovirus immediate-early enhancer, chicken β-actin promoter, chimera between introns from chicken β-actin and rabbit β-globin. pDRIVE CAG plasmid (Invivogen, San Diego, Calif.; having 100% sequence homology with the pCAGGS). The University of Pennsylvania considers CBA and CAGGS the same. | [63] |
| mini CAG (SV40 Intron) | 800 | Ubiquitous, +++ | [64] |
| Mini CAG no intron | 250 | chicken β-actin promoter, Ubiquitous, + | [57] |
| CBA/CB7 | 800 | Ubiquitous, ++ | [65] |
| smCBA | 953 | Ubiquitous, ? | [66] |
| CBh | 800 | CBA.MVM Ubiquitous, ++ | [54,67] |
| MeCP2 | 229 | ubiquitous | [68] |
| CMV | 800 | Ubiquitous, ++, prone to silencing | [54] |
| shCMV | 220 | Ubiquitous, ++ | [24] |
| CMVd2 | 52 | Low basal activity. Ubiquitous, Promega, + | [69] Cat.: pFN23A Halo Tag CMV d2 |
| core CMV | 30 | Not active without enhancers | [48] |
| SV40mini | 106 | SV40 minimal promoter | [48,49] |
| SCP3 | 81 | Super core promoter. (TATA box, Inr, MTE and DPE) | [48] |
| EF1-α | 2500 | Ubiquitous, ++ | [51,70] |
| PGK | 426 | Ubiquitous, ++ | [53] |
| UbC | 403 | Ubiquitous, ++ | [70] |
| Müller Glial Cells | Size (bp) | Origin, Cell Expression, Strength | References |
|---|---|---|---|
| CHX10 | 164 | Retinal progenitor cells | [98] |
| GFAP | 2600 | Müller glial cells, | [99,100] |
| GFAP | 2200 | Müller glial cells (Novartis) | [101] |
| GfaABC1D | 686 | Müller glial cells | [96,97] |
| HRSE-6xHRE-GfaABC1D | ~820 | Hypoxia-induced reactive MGC promoter. HRE is (A/G)CGT(G/C)C. HRSE from metallothionein II promoter (90 bps) | [97,102] |
| RLBP1 | 2789 | Müller glial cells | [24,89] |
| Short RLBP1 | 581 | Müller glial cells | [101] |
| Murine CD44 | 1775 | Müller glial cells | [24,95] |
| Murine shCD44 | 363 | Müller glial cells | [24,103] |
| ProB2 | 592 | Müller glial cells | [50] |
| Photoreceptor Cells | Size (bp) | Origin, Cell Expression, Strength | References |
| Mouse RHO | 1400 | Rod PRCs | [104] |
| Human RHO (rhodopsin) | 800 | Rod PRCs | [105] |
| Human RHO | 520 | Rod- PRCs | [24] |
| Mouse rod opsin mOp500 | 500 | Rod-PRCs −385/+86 | [106] |
| Mouse rod opsin | 221 | Rod-PRCs | [107] |
| Human Rhodopsin kinase (RHOK/GRK1) | 294 | Rod and cone PRCs. AY327580.1: bp 1793–2087 (−112 to +180). More efficient than IRBP in NHP for cone transduction | [24,91,108,109,110] |
| Human blue opsin HB570 | 570 | S-cone and subset of M-cones PRCs | [111] |
| Human blue opsin HB569 | 569 | blue cone opsin PRCs | [106,112] |
| PR0.5 | 496 | Red cone PRCs | [106] |
| PR1.7 | 1700 | Red cone PRCs | [106] |
| PR2.1 | 2100 | Red cone PRCs | [106] |
| 3LCR-PR0.5 | ~600 | Red cone PRCs | [106] |
| Mouse blue opsin (mBP500) | 500 | Mouse S opsin | [113] |
| Human interphotoreceptor retinoid binding protein (hIRBP) | 235 | Cone and rod PRCs X53044.1, bp 2603–2837 | [114] |
| IRBPe/GNAT2 | 500 | Cone PRCs | [115] |
| Mouse CAR/ARR3 | 500 | Cone PRCs, some rods, and RPE | [115] |
| Human CAR/ARR3 | 405–500 | Cone PRCs, some rods, and RPE cells | [115,116] |
| CAR/ARR3 | 215 | Cone PRC | [117] |
| Human red opsin | 2100 | Human red cone opsin | [118] |
| Human green red opsin (G1.7p) | 1700 | Cone PRCs. Core green opsin promoter including a mutation (0.5 kb) + Locus Control Region (LCR; 1.2 kb) upstream of the red opsin gene | [119,120,121] |
| Crx2kb | 2000 | Cone and rod PRCs | [122] |
| ProA1 | 2000 | cone PRCs | [50] |
| ProA4 | 2000 | cone PRCs | [50] |
| ProC1 | 731 | Cone and rod PRCs | [50] |
| ProA6,ProB5,ProC22,ProC32,ProD2,ProD3, ProD4,ProD5,ProD6 | 1229, 619 774, 814, 366, 691, 552, 321, 448 | rod PRCs | [50] |
| Synp161 | 150 | Mouse CD47 enhancer + SV40-mini promoter. Rod PRCs | [49] |
| Bipolar Cells | Size (bp) | Origin, Cell Expression, Strength | References |
| Mouse metabotropic glutamate receptor 6 (mGrm6) | 200 | On-bipolar cells | [98] |
| 4× mGRM6e+SV40 | 1000 | On-bipolar cells. 203 bp SV40 minimal promoter | [123] |
| Grm6e-Chx10-Cabp5 | 809 | 200 bp Grm6 + 164 bps Chx10 enhancer + 445 bp Cabp5 promoter. Wide overlapping bipolar expression | [98] |
| Grm6-SV40 | 400 | Grm6=mGluR6. 200 bp mGluR6 enhancer + SV40 promoter. On-Bipolar cells | [98] |
| Cabp5 | 445 | Bipolar cells | [98] |
| Chx10-SV40 | 364 | 164 bp Chx10 enhancer + 200 bp SV40 promoter. Bipolar cells and Müller glial cells | [98] |
| Grm6-mGluR500P | 700 | On-bipolar cells. | [124] |
| In4s-In3e- Grm6-mGluR500P | 1997 | 690 bp shortened Intron 4s + 807 bp Intron 3 + 500 bp mGluR500P | [124] |
| ProB4 | 1317 | Off-bipolar cells | [50] |
| Amacrine Cells | Size (bp) | Origin, Cell Expression, Strength | References |
| ProC2 | 964 | All amacrine cells + few MGCs | [50] |
| ProB1 | 394 | Amacrines with processes in one stratum | [50] |
| Horizontal Cells | Size (bp) | Origin, Cell expression, Strength | References |
| ProC3 | 694 | Some off-target in amacrine and ganglion cells | [50] |
| Retinal Ganglion Cells | Size (bp) | Origin, Cell expression, Strength | References |
| Syn1 | 495 | Off target amacrine, strength: ++ | [125] |
| Nefh | 2251 | Strength: +++ | [90] |
| hSNCGp | 948 | Human SNCG promoter (−785 to +163 region) | [126] |
| ProA3 | 2000 | Synthetic | [50] |
| Ple344 | 801 | Gene TUBB3. GCL and corneal nerves. ++ | [127] |
| Ple345 | 2693 | Gene NEFL. +++ (stronger than smCBA) | [127] |
| RPE | Size (bp) | Origin, Cell Expression, Strength | References |
| hRPE65p | 1383 | Chr1.68449936-68451318. RPE+ some PRC infection | [128] |
| NA65p | 1383 | Codon optimized hRPE65p+SV40 intron + Kozak seq, 150× more efficient than CBA and 300× more efficient than hRPE65p | [36] |
| VMD2 | 646 | NG_009033.1, bp 4870–5516 | [126,129] |
| Synpiii | 1317 | + SV40 mini promoter | [130] |
| Introns and PRE and Enhancers | Size | Description, Strength | References |
|---|---|---|---|
| CE (CMV early enhancer) | 431 | +++, 1.5–67× increase; −118/−522 TSS pCMVβ/5′CMV enhancer | [149] |
| IRBPe | 235 | human interphotoreceptor retinoid-binding protein proximal enhancer. Upstream nt −1619/−141 IRBP | [115] |
| metabotropic glutamate receptor 6 enhancer (Grm6e) | 200 | Grm6 proximal enhancer | [98] |
| Woodchuck Hepatitis Virus PRE (WPRE) | 600 | +++, 6–10× increase | [139,150] |
| Hepatitis B Virus PRE (HPRE) | 533 | +++, 6–10× increase | [150] |
| WPRE3 | 247 | ++, 6× increase | [139] |
| MVM | 67–97 | +++, minute virus of mice, 10× increase | [143] |
| chCMV.HBB2 | ~506 | Chimeric CMV (146 bp) + human β-globulin intron 2 (340 bp) + exon 3 20 bp incl SA/SD | [151] |
| Hybrid adenovirus SD#/IgG Sa* | 230 | +++, pAdβ, 2× increase to synthetic polyA | [149] |
| SV40 late SD#/Sa* (19S/16S) | 180 | +, pCMVβ (Promega; 1.6× increase) | [149] |
| Modified SV40 SD#/Sa* | 157 | modSV40 SA/SD= modified SV40 splice acceptor/donor intron, 157 bp in length, nucleotides 502–561 and 1410–1497 of SV40 genomic sequence (NC_001669.1) + connecting sequence CGGATCCGG between two fragments. | [101,152] |
| Mini SV40 SD#/Sa* | 100 | Mini SV40 SD#/Sa* intron | [43,153,154] |
| Human β -globin intron 2 SD#/Sa* | 875 | 0.5–86-fold increase. pZac2.1 | [139,155,156,157] |
| F.IX truncated intron1 | 300 | +, human factor IX (100×) | [143,158] |
| Miscellaneous | Size | Description | References |
| 2A | 75 | Self-cleaving linker | [159] |
| internal ribosomal entry site (IRES) | 600 | Ubiquitous. Placed between two genes. The second gene is transcribed without a promoter (at a lower expression compared to the first gene) | [160] |
| SPTP | 154 | Synthetic polyA signal/transcriptional pause site frp, pGL4.25 | [161] |
| PolII miR-155 | ~500 | Block-iT PolII miR vector system based on miR-155 expressing artificial miRNAs engineered to a target sequence resulting in target cleavage | [162] |
| shRNA-YB1 | N/A | 7-to-45 fold AAV production increase in physical titer | [163] |
| MIP backbone | N/A | mini-intronic plasmid (MIP) backbones for AAV production increased transgene expression by 40–100 fold in vivo | [144] |
| R6K | 545 | + (~40×),pUC + prokaryotic RNA-OUT antibiotic-free, minicircle AAVs | [144] |
| OIPR | 1300 | + (~40×),pUC + prokaryotic RNA-OUT antibiotic-free, minicircle AAVs | [144] |
| Shorter OIPR | 500 | + (~5×),pUC + prokaryotic RNA-OUT antibiotic-free, minicircle AAVs | [144] |
| Polyadenylation | Size | Description, Strength | References |
|---|---|---|---|
| SV40 late | 135 | +++ | [139] |
| 2× SV40 late | 100 | ++/+++ | [169] |
| bGHpolyA | 250 | ++ | [149] |
| 2× sNRP1 | 34 | +/++ | [167] |
| Rabbit gbpA | 56 | Rabbit β-globin | [149] |
| spA | 49 | +/++ (7× lower than bGHpolyA, 3× lower than SV40 late) | [139,149] |
| hGHpolyA | 624 | + | [41,170,171] |
| 1× sNRP1 | 17 | + | [167] |
| HSV TK poly(A) | 48 | herpes simplex virus (HSV) thymidine kinase (TK) polyadenylation signal. Generally used for NeoR and KanR genes | [172] |
| Adenovirus (L3) USE | 21 | + | [169] |
| Inducible Promoters | Size (bp) | Origin, Cell Expression, Strength | References |
|---|---|---|---|
| MT-1 | 13,200 | Zinc, cadmium or copper-inducible sheep metallothionine-Ia promoter | [174] |
| MMTV LTR | 792 | dexamethasone (Dex)-inducible mouse mammary tumor virus. Active when glucocorticoids or progestins present | [175,176] |
| Ptet | 270 | tetracycline On or Off system promoters (Ptet). 6× mutated TRE (~200 bp) core CMV (~40 bps) | [177] |
| T7lac | 42 | T7 bacteriophage promoter (17 bp) requires T7 RNA polymerase and lac operator (25 bp). Induces expression by IPTG | [180] |
| Riboswitches | ~100 | ligand-sensing aptamer, a communication module (linker), and an effector domain (ribozyme) | [173,178,179] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Buck, T.M.; Wijnholds, J. Recombinant Adeno-Associated Viral Vectors (rAAV)-Vector Elements in Ocular Gene Therapy Clinical Trials and Transgene Expression and Bioactivity Assays. Int. J. Mol. Sci. 2020, 21, 4197. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21124197
Buck TM, Wijnholds J. Recombinant Adeno-Associated Viral Vectors (rAAV)-Vector Elements in Ocular Gene Therapy Clinical Trials and Transgene Expression and Bioactivity Assays. International Journal of Molecular Sciences. 2020; 21(12):4197. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21124197
Chicago/Turabian StyleBuck, Thilo M., and Jan Wijnholds. 2020. "Recombinant Adeno-Associated Viral Vectors (rAAV)-Vector Elements in Ocular Gene Therapy Clinical Trials and Transgene Expression and Bioactivity Assays" International Journal of Molecular Sciences 21, no. 12: 4197. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21124197