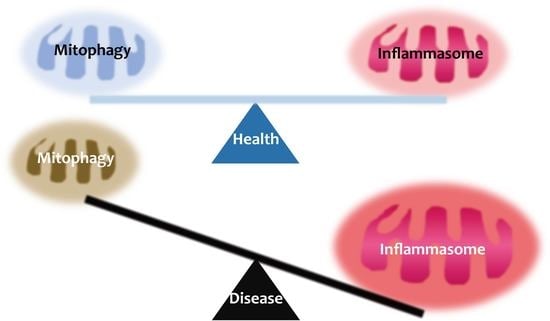
Inflammasome and Mitophagy Connection in Health and Disease
Abstract
:1. Introduction
2. Overview of Inflammasomes
2.1. NLRP3 Inflammasome
2.2. AIM2 Inflammasome
2.3. NAIP/NLRC4 Inflammasome
3. Overview of Mitophagy
3.1. PINK1/Parkin-Mediated Mitophagy
3.2. The Mitophagy Receptor-Mediated Pathway
3.3. Lipid-Mediated Mitophagy: Cardiolipin
4. Mitophagy Crosstalk with the Inflammasome
4.1. Mitochondrial Reactive Oxygen Species (ROS) Generation and Translocation of Mitochondrial DNA into the Cytosol
4.2. Multiple Regulators That Connect between Inflammasome and Mitophagy
5. Crosstalk between Inflammasomes and Mitophagy during Infection
5.1. The Inflammasome-Mitophagy Axis in Bacterial Infections
5.2. The Inflammasome-Mitophagy Axis in Viral Infections
6. Altered Crosstalk between Inflammasome and Mitophagy in the Context of Human Disease
6.1. Metabolic and Cardiovascular Diseases
6.2. Neuronal and Nephrotic Inflammation, and Sepsis
6.3. Cancers
7. The Pharmacological Regulation of Inflammasome-Mitophagy Axis to Control Human Disease
7.1. Pharmacological Regulation in Models of Metabolic and Cardiovascular Diseases
7.2. Pharmacological Regulation in Models of Neurological and Nephrotic Diseases
7.3. Pharmacological Regulation in Models of Inflammatory Disease: Sepsis and Colitis
7.4. Pharmacological Regulation Targeting Sirtuins Nuclear Receptor Rev-erbα
8. Conclusions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AIM2 | Absent in melanoma 2 |
| AMPK | AMP-activated protein kinase |
| AMPK | 5′-AMP-activated protein kinase |
| ASC | Apoptosis-associated speck-like protein containing a CARD |
| Atg | Autophagy-related gene |
| BCCAo | Bilateral common carotid artery occlusion |
| BMDM | Bone marrow-derived macrophages |
| CAC | Colitis-associated cancer |
| CCH | Chronic cerebral hypoperfusion |
| CK2 | Casein kinase 2 |
| CLP | Cecal ligation and puncture |
| DAMP | Damage associated molecular pattern |
| dsDNA | Double-stranded DNA |
| DSS | Dextran sulfate sodium |
| eIF2α | Eukaryotic initiation factor 2 |
| FUNDC1 | FUN14 domain containing 1 |
| GABARAP | Gamma-aminobutyric acid receptor-associated protein |
| GLP-1 | Glucagon like peptide-1 |
| HepG2 | Human hepatocellular carcinoma cell line |
| HMGB1 | High mobility group box 1 |
| IL | Interleukin |
| IMM | Inner mitochondrial membrane |
| LPS | Lipopolysaccharide |
| MFN2 | Mitofusin 2 |
| mtDNA | Mitochondrial DNA |
| mTOR | Mammalian target of rapamycin |
| mtROS | Mitochondrial reactive oxygen species |
| NAIP | NLR family, apoptosis inhibitory protein |
| NEK7 | NIMA related kinase 7 |
| NF-κB | Nuclear factor kappa-light-chain-enhancer of activated B cells |
| NIX | NIP3-like protein X |
| NLRC4 | NLR family CARD domain containing 4 |
| NLRP3 | NOD-, LRR- and pyrin domain-containing protein 3 |
| NR1D1 | Nuclear receptor subfamily 1, group D, member 1 |
| OMM | Outer mitochondrial membrane |
| OPTN | Optineurin |
| ox-LDL | Oxidized low-density lipoprotein |
| P2X7 | P2X purinoceptor 7 |
| PA | Palmitic acid |
| PAMP | Pathogen associated molecular pattern |
| PBMC | Peripheral blood mononuclear cell |
| PGAM5 | PGAM family member 5 |
| PHB2 | Prohibitin 2 |
| PINK1 | PTEN Induced Kinase 1 |
| PKC | Protein kinase C |
| PRR | Pattern recognition receptor |
| RIPK2 | Receptor-interacting serine/threonine-protein kinase 2 |
| ROS | Reactive oxygen species |
| SAH | Subarachnoid hemorrhage |
| SESN2 | Sestrin 2 |
| SQSTM1 | Sequestosome 1 |
| T2DM | Type 2 diabetes mellitus |
| TLR | Toll-like receptor |
| TNF | Tumor necrosis factor |
| TNFR | Tumor necrosis factor receptor |
| UC | Ulcerative colitis |
| ULK1 | Unc-51 like kinase 1 |
References
- Zhong, Z.; Sanchez-Lopez, E.; Karin, M. Autophagy, NLRP3 inflammasome and auto-inflammatory/immune diseases. Clin. Exp. Rheumatol. 2016, 34, 12–16. [Google Scholar]
- Jo, E.K.; Kim, J.K.; Shin, D.M.; Sasakawa, C. Molecular mechanisms regulating NLRP3 inflammasome activation. Cell. Mol. Immunol. 2016, 13, 148–159. [Google Scholar] [CrossRef] [Green Version]
- Krakauer, T. Inflammasomes, Autophagy, and Cell Death: The Trinity of Innate Host Defense against Intracellular Bacteria. Mediat. Inflamm. 2019, 2019, 2471215. [Google Scholar] [CrossRef] [Green Version]
- Man, S.M.; Karki, R.; Kanneganti, T.D. Molecular mechanisms and functions of pyroptosis, inflammatory caspases and inflammasomes in infectious diseases. Immunol. Rev. 2017, 277, 61–75. [Google Scholar] [CrossRef] [Green Version]
- Jorgensen, I.; Miao, E.A. Pyroptotic cell death defends against intracellular pathogens. Immunol. Rev. 2015, 265, 130–142. [Google Scholar] [CrossRef]
- Spel, L.; Martinon, F. Inflammasomes contributing to inflammation in arthritis. Immunol. Rev. 2020, 294, 48–62. [Google Scholar] [CrossRef] [Green Version]
- Liang, F.; Zhang, F.; Zhang, L.; Wei, W. The advances in pyroptosis initiated by inflammasome in inflammatory and immune diseases. Inflamm. Res. 2020, 69, 159–166. [Google Scholar] [CrossRef]
- Lu, F.; Lan, Z.; Xin, Z.; He, C.; Guo, Z.; Xia, X.; Hu, T. Emerging insights into molecular mechanisms underlying pyroptosis and functions of inflammasomes in diseases. J. Cell. Physiol. 2020, 235, 3207–3221. [Google Scholar] [CrossRef]
- Khaminets, A.; Behl, C.; Dikic, I. Ubiquitin-Dependent And Independent Signals In Selective Autophagy. Trends Cell Biol. 2016, 26, 6–16. [Google Scholar] [CrossRef]
- Gustafsson, A.B.; Dorn, G.W., 2nd. Evolving and Expanding the Roles of Mitophagy as a Homeostatic and Pathogenic Process. Physiol. Rev. 2019, 99, 853–892. [Google Scholar] [CrossRef]
- Eiyama, A.; Okamoto, K. PINK1/Parkin-mediated mitophagy in mammalian cells. Curr. Opin. Cell. Biol. 2015, 33, 95–101. [Google Scholar] [CrossRef]
- Wei, H.; Liu, L.; Chen, Q. Selective removal of mitochondria via mitophagy: Distinct pathways for different mitochondrial stresses. Biochim. Biophys. Acta 2015, 1853, 2784–2790. [Google Scholar] [CrossRef] [Green Version]
- A, A.M.; Ameenudeen, S.; Kumar, A.; Hemalatha, S.; Ahmed, N.; Ali, N.; AlAsmari, A.F.; Aashique, M.; Waseem, M. Emerging Role of Mitophagy in Inflammatory Diseases: Cellular and Molecular Episodes. Curr. Pharm. Des. 2020, 26, 485–491. [Google Scholar] [CrossRef]
- Fan, P.; Xie, X.H.; Chen, C.H.; Peng, X.; Zhang, P.; Yang, C.; Wang, Y.T. Molecular Regulation Mechanisms and Interactions Between Reactive Oxygen Species and Mitophagy. DNA Cell Biol 2019, 38, 10–22. [Google Scholar] [CrossRef]
- Zhao, Y.; Huang, S.; Liu, J.; Wu, X.; Zhou, S.; Dai, K.; Kou, Y. Mitophagy Contributes to the Pathogenesis of Inflammatory Diseases. Inflammation 2018, 41, 1590–1600. [Google Scholar] [CrossRef]
- Harris, J.; Deen, N.; Zamani, S.; Hasnat, M.A. Mitophagy and the release of inflammatory cytokines. Mitochondrion 2018, 41, 2–8. [Google Scholar] [CrossRef]
- Kim, M.J.; Yoon, J.H.; Ryu, J.H. Mitophagy: A balance regulator of NLRP3 inflammasome activation. BMB Rep. 2016, 49, 529–535. [Google Scholar] [CrossRef]
- Hamacher-Brady, A.; Brady, N.R. Mitophagy programs: Mechanisms and physiological implications of mitochondrial targeting by autophagy. Cell. Mol. Life Sci. 2016, 73, 775–795. [Google Scholar] [CrossRef] [Green Version]
- Ashrafi, G.; Schwarz, T.L. The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death Differ. 2013, 20, 31–42. [Google Scholar] [CrossRef] [Green Version]
- Jang, J.H.; Shin, H.W.; Lee, J.M.; Lee, H.W.; Kim, E.C.; Park, S.H. An Overview of Pathogen Recognition Receptors for Innate Immunity in Dental Pulp. Mediat. Inflamm. 2015, 2015, 794143. [Google Scholar] [CrossRef] [Green Version]
- Kelley, N.; Jeltema, D.; Duan, Y.; He, Y. The NLRP3 Inflammasome: An Overview of Mechanisms of Activation and Regulation. Int. J. Mol. Sci. 2019, 20, 3328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Howrylak, J.A.; Nakahira, K. Inflammasomes: Key Mediators of Lung Immunity. Annu. Rev. Physiol. 2017, 79, 471–494. [Google Scholar] [CrossRef]
- Wang, B.; Tian, Y.; Yin, Q. AIM2 Inflammasome Assembly and Signaling. Adv. Exp. Med. Biol. 2019, 1172, 143–155. [Google Scholar] [CrossRef] [PubMed]
- Andrade, W.A.; Zamboni, D.S. NLRC4 biology in immunity and inflammation. J. Leukoc. Biol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Hayward, J.A.; Mathur, A.; Ngo, C.; Man, S.M. Cytosolic Recognition of Microbes and Pathogens: Inflammasomes in Action. Microbiol. Mol. Biol. Rev. 2018, 82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guey, B.; Bodnar, M.; Manie, S.N.; Tardivel, A.; Petrilli, V. Caspase-1 autoproteolysis is differentially required for NLRP1b and NLRP3 inflammasome function. Proc. Natl. Acad. Sci. USA 2014, 111, 17254–17259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Opdenbosch, N.; Gurung, P.; Vande Walle, L.; Fossoul, A.; Kanneganti, T.D.; Lamkanfi, M. Activation of the NLRP1b inflammasome independently of ASC-mediated caspase-1 autoproteolysis and speck formation. Nat. Commun. 2014, 5, 3209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loeven, N.A.; Medici, N.P.; Bliska, J.B. The pyrin inflammasome in host-microbe interactions. Curr. Opin. Microbiol. 2020, 54, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Manukyan, G.; Aminov, R. Update on Pyrin Functions and Mechanisms of Familial Mediterranean Fever. Front. Microbiol. 2016, 7, 456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prochnicki, T.; Latz, E. Inflammasomes on the Crossroads of Innate Immune Recognition and Metabolic Control. Cell Metab. 2017, 26, 71–93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christgen, S.; Place, D.E.; Kanneganti, T.D. Toward targeting inflammasomes: Insights into their regulation and activation. Cell Res. 2020, 30, 315–327. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.; Suh, G.Y.; Ryter, S.W.; Choi, A.M. Regulation and Function of the Nucleotide Binding Domain Leucine-Rich Repeat-Containing Receptor, Pyrin Domain-Containing-3 Inflammasome in Lung Disease. Am. J. Respir. Cell Mol. Biol. 2016, 54, 151–160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strowig, T.; Henao-Mejia, J.; Elinav, E.; Flavell, R. Inflammasomes in health and disease. Nature 2012, 481, 278–286. [Google Scholar] [CrossRef] [PubMed]
- Yen, W.C.; Wu, Y.H.; Wu, C.C.; Lin, H.R.; Stern, A.; Chen, S.H.; Shu, J.C.; Tsun-Yee Chiu, D. Impaired inflammasome activation and bacterial clearance in G6PD deficiency due to defective NOX/p38 MAPK/AP-1 redox signaling. Redox Biol. 2020, 28, 101363. [Google Scholar] [CrossRef]
- Chen, H.; Jiang, Z. The essential adaptors of innate immune signaling. Protein Cell 2013, 4, 27–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elliott, E.I.; Sutterwala, F.S. Initiation and perpetuation of NLRP3 inflammasome activation and assembly. Immunol. Rev. 2015, 265, 35–52. [Google Scholar] [CrossRef] [Green Version]
- Broz, P.; Dixit, V.M. Inflammasomes: Mechanism of assembly, regulation and signalling. Nat. Rev. Immunol. 2016, 16, 407–420. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, H.; Kouadir, M.; Song, H.; Shi, F. Recent advances in the mechanisms of NLRP3 inflammasome activation and its inhibitors. Cell Death Dis. 2019, 10, 128. [Google Scholar] [CrossRef] [Green Version]
- Chen, K.W.; Demarco, B.; Broz, P. Pannexin-1 promotes NLRP3 activation during apoptosis but is dispensable for canonical or noncanonical inflammasome activation. Eur. J. Immunol. 2020, 50, 170–177. [Google Scholar] [CrossRef]
- Chen, K.W.; Demarco, B.; Heilig, R.; Shkarina, K.; Boettcher, A.; Farady, C.J.; Pelczar, P.; Broz, P. Extrinsic and intrinsic apoptosis activate pannexin-1 to drive NLRP3 inflammasome assembly. EMBO J. 2019, 38. [Google Scholar] [CrossRef]
- Yu, J.W.; Lee, M.S. Mitochondria and the NLRP3 inflammasome: Physiological and pathological relevance. Arch. Pharm. Res. 2016, 39, 1503–1518. [Google Scholar] [CrossRef] [PubMed]
- Abais, J.M.; Xia, M.; Zhang, Y.; Boini, K.M.; Li, P.L. Redox regulation of NLRP3 inflammasomes: ROS as trigger or effector? Antioxid. Redox Signal. 2015, 22, 1111–1129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tschopp, J. Mitochondria: Sovereign of inflammation? Eur. J. Immunol. 2011, 41, 1196–1202. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Zhang, D.; Hu, D.; Zhou, X.; Zhou, Y. The role of mitochondria in NLRP3 inflammasome activation. Mol. Immunol. 2018, 103, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Iyer, S.S.; He, Q.; Janczy, J.R.; Elliott, E.I.; Zhong, Z.; Olivier, A.K.; Sadler, J.J.; Knepper-Adrian, V.; Han, R.; Qiao, L.; et al. Mitochondrial cardiolipin is required for Nlrp3 inflammasome activation. Immunity 2013, 39, 311–323. [Google Scholar] [CrossRef] [Green Version]
- Mohanty, A.; Tiwari-Pandey, R.; Pandey, N.R. Mitochondria: The indispensable players in innate immunity and guardians of the inflammatory response. J. Cell Commun. Signal. 2019, 13, 303–318. [Google Scholar] [CrossRef]
- Park, S.; Juliana, C.; Hong, S.; Datta, P.; Hwang, I.; Fernandes-Alnemri, T.; Yu, J.W.; Alnemri, E.S. The mitochondrial antiviral protein MAVS associates with NLRP3 and regulates its inflammasome activity. J. Immunol. 2013, 191, 4358–4366. [Google Scholar] [CrossRef] [Green Version]
- Subramanian, N.; Natarajan, K.; Clatworthy, M.R.; Wang, Z.; Germain, R.N. The adaptor MAVS promotes NLRP3 mitochondrial localization and inflammasome activation. Cell 2013, 153, 348–361. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Zeng, M.Y.; Yang, D.; Motro, B.; Nunez, G. NEK7 is an essential mediator of NLRP3 activation downstream of potassium efflux. Nature 2016, 530, 354–357. [Google Scholar] [CrossRef] [Green Version]
- Shi, H.; Wang, Y.; Li, X.; Zhan, X.; Tang, M.; Fina, M.; Su, L.; Pratt, D.; Bu, C.H.; Hildebrand, S.; et al. NLRP3 activation and mitosis are mutually exclusive events coordinated by NEK7, a new inflammasome component. Nat. Immunol. 2016, 17, 250–258. [Google Scholar] [CrossRef]
- Schmid-Burgk, J.L.; Chauhan, D.; Schmidt, T.; Ebert, T.S.; Reinhardt, J.; Endl, E.; Hornung, V. A Genome-wide CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) Screen Identifies NEK7 as an Essential Component of NLRP3 Inflammasome Activation. J. Biol. Chem. 2016, 291, 103–109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yi, Y.S. Caspase-11 non-canonical inflammasome: A critical sensor of intracellular lipopolysaccharide in macrophage-mediated inflammatory responses. Immunology 2017, 152, 207–217. [Google Scholar] [CrossRef]
- Casson, C.N.; Copenhaver, A.M.; Zwack, E.E.; Nguyen, H.T.; Strowig, T.; Javdan, B.; Bradley, W.P.; Fung, T.C.; Flavell, R.A.; Brodsky, I.E.; et al. Caspase-11 activation in response to bacterial secretion systems that access the host cytosol. PLoS Pathog. 2013, 9, e1003400. [Google Scholar] [CrossRef]
- He, W.T.; Wan, H.; Hu, L.; Chen, P.; Wang, X.; Huang, Z.; Yang, Z.H.; Zhong, C.Q.; Han, J. Gasdermin D is an executor of pyroptosis and required for interleukin-1beta secretion. Cell Res. 2015, 25, 1285–1298. [Google Scholar] [CrossRef]
- Shi, J.; Zhao, Y.; Wang, K.; Shi, X.; Wang, Y.; Huang, H.; Zhuang, Y.; Cai, T.; Wang, F.; Shao, F. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 2015, 526, 660–665. [Google Scholar] [CrossRef]
- Kuriakose, T.; Kanneganti, T.D. Regulation and functions of NLRP3 inflammasome during influenza virus infection. Mol. Immunol. 2017, 86, 56–64. [Google Scholar] [CrossRef]
- Rabes, A.; Suttorp, N.; Opitz, B. Inflammasomes in Pneumococcal Infection: Innate Immune Sensing and Bacterial Evasion Strategies. Curr. Top. Microbiol. Immunol. 2016, 397, 215–227. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.J.; Jo, E.K. NLRP3 inflammasome and host protection against bacterial infection. J. Korean Med. Sci. 2013, 28, 1415–1423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franchi, L.; Warner, N.; Viani, K.; Nunez, G. Function of Nod-like receptors in microbial recognition and host defense. Immunol. Rev. 2009, 227, 106–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christgen, S.; Kanneganti, T.D. Inflammasomes and the fine line between defense and disease. Curr. Opin. Immunol. 2020, 62, 39–44. [Google Scholar] [CrossRef]
- Awad, F.; Assrawi, E.; Louvrier, C.; Jumeau, C.; Georgin-Lavialle, S.; Grateau, G.; Amselem, S.; Giurgea, I.; Karabina, S.A. Inflammasome biology, molecular pathology and therapeutic implications. Pharmacol. Ther. 2018, 187, 133–149. [Google Scholar] [CrossRef]
- Kesavardhana, S.; Kanneganti, T.D. Mechanisms governing inflammasome activation, assembly and pyroptosis induction. Int. Immunol. 2017, 29, 201–210. [Google Scholar] [CrossRef]
- Burckstummer, T.; Baumann, C.; Bluml, S.; Dixit, E.; Durnberger, G.; Jahn, H.; Planyavsky, M.; Bilban, M.; Colinge, J.; Bennett, K.L.; et al. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat. Immunol. 2009, 10, 266–272. [Google Scholar] [CrossRef]
- Fernandes-Alnemri, T.; Yu, J.W.; Datta, P.; Wu, J.; Alnemri, E.S. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature 2009, 458, 509–513. [Google Scholar] [CrossRef] [Green Version]
- Hornung, V.; Ablasser, A.; Charrel-Dennis, M.; Bauernfeind, F.; Horvath, G.; Caffrey, D.R.; Latz, E.; Fitzgerald, K.A. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature 2009, 458, 514–518. [Google Scholar] [CrossRef] [Green Version]
- Roberts, T.L.; Idris, A.; Dunn, J.A.; Kelly, G.M.; Burnton, C.M.; Hodgson, S.; Hardy, L.L.; Garceau, V.; Sweet, M.J.; Ross, I.L.; et al. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science 2009, 323, 1057–1060. [Google Scholar] [CrossRef] [Green Version]
- Marim, F.M.; Franco, M.M.C.; Gomes, M.T.R.; Miraglia, M.C.; Giambartolomei, G.H.; Oliveira, S.C. The role of NLRP3 and AIM2 in inflammasome activation during Brucella abortus infection. Semin Immunopathol. 2017, 39, 215–223. [Google Scholar] [CrossRef] [Green Version]
- Sauer, J.D.; Witte, C.E.; Zemansky, J.; Hanson, B.; Lauer, P.; Portnoy, D.A. Listeria monocytogenes triggers AIM2-mediated pyroptosis upon infrequent bacteriolysis in the macrophage cytosol. Cell Host Microbe 2010, 7, 412–419. [Google Scholar] [CrossRef] [Green Version]
- Fernandes-Alnemri, T.; Yu, J.W.; Juliana, C.; Solorzano, L.; Kang, S.; Wu, J.; Datta, P.; McCormick, M.; Huang, L.; McDermott, E.; et al. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nat. Immunol. 2010, 11, 385–393. [Google Scholar] [CrossRef] [Green Version]
- Rathinam, V.A.; Jiang, Z.; Waggoner, S.N.; Sharma, S.; Cole, L.E.; Waggoner, L.; Vanaja, S.K.; Monks, B.G.; Ganesan, S.; Latz, E.; et al. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat. Immunol. 2010, 11, 395–402. [Google Scholar] [CrossRef] [Green Version]
- Man, S.M.; Zhu, Q.; Zhu, L.; Liu, Z.; Karki, R.; Malik, A.; Sharma, D.; Li, L.; Malireddi, R.K.; Gurung, P.; et al. Critical Role for the DNA Sensor AIM2 in Stem Cell Proliferation and Cancer. Cell 2015, 162, 45–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dombrowski, Y.; Peric, M.; Koglin, S.; Kammerbauer, C.; Goss, C.; Anz, D.; Simanski, M.; Glaser, R.; Harder, J.; Hornung, V.; et al. Cytosolic DNA triggers inflammasome activation in keratinocytes in psoriatic lesions. Sci. Transl. Med. 2011, 3, 82ra38. [Google Scholar] [CrossRef] [Green Version]
- Javierre, B.M.; Fernandez, A.F.; Richter, J.; Al-Shahrour, F.; Martin-Subero, J.I.; Rodriguez-Ubreva, J.; Berdasco, M.; Fraga, M.F.; O’Hanlon, T.P.; Rider, L.G.; et al. Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Res. 2010, 20, 170–179. [Google Scholar] [CrossRef] [Green Version]
- Bae, J.H.; Jo, S.I.; Kim, S.J.; Lee, J.M.; Jeong, J.H.; Kang, J.S.; Cho, N.J.; Kim, S.S.; Lee, E.Y.; Moon, J.S. Circulating Cell-Free mtDNA Contributes to AIM2 Inflammasome-Mediated Chronic Inflammation in Patients with Type 2 Diabetes. Cells 2019, 8, 328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bauer, R.; Rauch, I. The NAIP/NLRC4 inflammasome in infection and pathology. Mol. Aspects Med. 2020. [Google Scholar] [CrossRef] [PubMed]
- Haloupek, N.; Grob, P.; Tenthorey, J.; Vance, R.E.; Nogales, E. Cryo-EM studies of NAIP-NLRC4 inflammasomes. Methods Enzymol. 2019, 625, 177–204. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Zhou, Q.; Zhang, C.; Fan, S.; Cheng, W.; Zhao, Y.; Shao, F.; Wang, H.W.; Sui, S.F.; Chai, J. Structural and biochemical basis for induced self-propagation of NLRC4. Science 2015, 350, 399–404. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Chen, S.; Ruan, J.; Wu, J.; Tong, A.B.; Yin, Q.; Li, Y.; David, L.; Lu, A.; Wang, W.L.; et al. Cryo-EM structure of the activated NAIP2-NLRC4 inflammasome reveals nucleated polymerization. Science 2015, 350, 404–409. [Google Scholar] [CrossRef] [Green Version]
- Moghaddas, F.; Zeng, P.; Zhang, Y.; Schutzle, H.; Brenner, S.; Hofmann, S.R.; Berner, R.; Zhao, Y.; Lu, B.; Chen, X.; et al. Autoinflammatory mutation in NLRC4 reveals a leucine-rich repeat (LRR)-LRR oligomerization interface. J. Allergy Clin. Immunol. 2018, 142, 1956–1967e1956. [Google Scholar] [CrossRef] [Green Version]
- Hausmann, A.; Bock, D.; Geiser, P.; Berthold, D.L.; Fattinger, S.A.; Furter, M.; Bouman, J.A.; Barthel-Scherrer, M.; Lang, C.M.; Bakkeren, E.; et al. Intestinal epithelial NAIP/NLRC4 restricts systemic dissemination of the adapted pathogen Salmonella Typhimurium due to site-specific bacterial PAMP expression. Mucosal Immunol. 2020, 13, 530–544. [Google Scholar] [CrossRef] [Green Version]
- West, T.E.; Myers, N.D.; Chantratita, N.; Chierakul, W.; Limmathurotsakul, D.; Wuthiekanun, V.; Miao, E.A.; Hajjar, A.M.; Peacock, S.J.; Liggitt, H.D.; et al. NLRC4 and TLR5 each contribute to host defense in respiratory melioidosis. PLoS Negl. Trop. Dis. 2014, 8, e3178. [Google Scholar] [CrossRef]
- Tenthorey, J.L.; Chavez, R.A.; Thompson, T.W.; Deets, K.A.; Vance, R.E.; Rauch, I. NLRC4 inflammasome activation is NLRP3- and phosphorylation-independent during infection and does not protect from melanoma. J. Exp. Med. 2020, 217. [Google Scholar] [CrossRef] [PubMed]
- Xian, H.; Yang, Q.; Xiao, L.; Shen, H.M.; Liou, Y.C. STX17 dynamically regulated by Fis1 induces mitophagy via hierarchical macroautophagic mechanism. Nat. Commun. 2019, 10, 2059. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhujabal, Z.; Birgisdottir, A.B.; Sjottem, E.; Brenne, H.B.; Overvatn, A.; Habisov, S.; Kirkin, V.; Lamark, T.; Johansen, T. FKBP8 recruits LC3A to mediate Parkin-independent mitophagy. EMBO Rep. 2017, 18, 947–961. [Google Scholar] [CrossRef] [PubMed]
- Szargel, R.; Shani, V.; Abd Elghani, F.; Mekies, L.N.; Liani, E.; Rott, R.; Engelender, S. The PINK1, synphilin-1 and SIAH-1 complex constitutes a novel mitophagy pathway. Hum. Mol. Genet. 2016, 25, 3476–3490. [Google Scholar] [CrossRef] [Green Version]
- Ambivero, C.T.; Cilenti, L.; Main, S.; Zervos, A.S. Mulan E3 ubiquitin ligase interacts with multiple E2 conjugating enzymes and participates in mitophagy by recruiting GABARAP. Cell. Signal. 2014, 26, 2921–2929. [Google Scholar] [CrossRef]
- Ding, W.X.; Yin, X.M. Mitophagy: Mechanisms, pathophysiological roles, and analysis. Biol. Chem. 2012, 393, 547–564. [Google Scholar] [CrossRef] [Green Version]
- Novak, I. Mitophagy: A complex mechanism of mitochondrial removal. Antioxid. Redox Signal. 2012, 17, 794–802. [Google Scholar] [CrossRef]
- Nguyen, T.N.; Padman, B.S.; Lazarou, M. Deciphering the Molecular Signals of PINK1/Parkin Mitophagy. Trends Cell Biol. 2016, 26, 733–744. [Google Scholar] [CrossRef]
- Barazzuol, L.; Giamogante, F.; Brini, M.; Cali, T. PINK1/Parkin Mediated Mitophagy, Ca(2+) Signalling, and ER-Mitochondria Contacts in Parkinson’s Disease. Int. J. Mol. Sci. 2020, 21, 1772. [Google Scholar] [CrossRef] [Green Version]
- Bingol, B.; Sheng, M. Mechanisms of mitophagy: PINK1, Parkin, USP30 and beyond. Free Radic. Biol. Med. 2016, 100, 210–222. [Google Scholar] [CrossRef] [PubMed]
- Rodger, C.E.; McWilliams, T.G.; Ganley, I.G. Mammalian mitophagy—From in vitro molecules to in vivo models. FEBS J. 2018, 285, 1185–1202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McWilliams, T.G.; Muqit, M.M. PINK1 and Parkin: Emerging themes in mitochondrial homeostasis. Curr. Opin. Cell Biol. 2017, 45, 83–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirkin, V.; McEwan, D.G.; Novak, I.; Dikic, I. A role for ubiquitin in selective autophagy. Mol. Cell 2009, 34, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Kirkin, V.; Rogov, V.V. A Diversity of Selective Autophagy Receptors Determines the Specificity of the Autophagy Pathway. Mol. Cell 2019, 76, 268–285. [Google Scholar] [CrossRef]
- Lamark, T.; Svenning, S.; Johansen, T. Regulation of selective autophagy: The p62/SQSTM1 paradigm. Essays Biochem. 2017, 61, 609–624. [Google Scholar] [CrossRef]
- Heo, J.M.; Ordureau, A.; Paulo, J.A.; Rinehart, J.; Harper, J.W. The PINK1-PARKIN Mitochondrial Ubiquitylation Pathway Drives a Program of OPTN/NDP52 Recruitment and TBK1 Activation to Promote Mitophagy. Mol. Cell 2015, 60, 7–20. [Google Scholar] [CrossRef] [Green Version]
- Lazarou, M.; Sliter, D.A.; Kane, L.A.; Sarraf, S.A.; Wang, C.; Burman, J.L.; Sideris, D.P.; Fogel, A.I.; Youle, R.J. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature 2015, 524, 309–314. [Google Scholar] [CrossRef] [Green Version]
- Wei, Y.; Chiang, W.C.; Sumpter, R., Jr.; Mishra, P.; Levine, B. Prohibitin 2 Is an Inner Mitochondrial Membrane Mitophagy Receptor. Cell 2017, 168, 224–238 e210. [Google Scholar] [CrossRef] [Green Version]
- Yan, C.; Gong, L.; Chen, L.; Xu, M.; Abou-Hamdan, H.; Tang, M.; Desaubry, L.; Song, Z. PHB2 (prohibitin 2) promotes PINK1-PRKN/Parkin-dependent mitophagy by the PARL-PGAM5-PINK1 axis. Autophagy 2020, 16, 419–434. [Google Scholar] [CrossRef]
- Shi, R.Y.; Zhu, S.H.; Li, V.; Gibson, S.B.; Xu, X.S.; Kong, J.M. BNIP3 interacting with LC3 triggers excessive mitophagy in delayed neuronal death in stroke. CNS Neurosci. Ther. 2014, 20, 1045–1055. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Ney, P.A. Role of BNIP3 and NIX in cell death, autophagy, and mitophagy. Cell Death Differ. 2009, 16, 939–946. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Sakakibara, K.; Chen, Q.; Okamoto, K. Receptor-mediated mitophagy in yeast and mammalian systems. Cell Res. 2014, 24, 787–795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsushima, M.; Fujiwara, T.; Takahashi, E.; Minaguchi, T.; Eguchi, Y.; Tsujimoto, Y.; Suzumori, K.; Nakamura, Y. Isolation, mapping, and functional analysis of a novel human cDNA (BNIP3L) encoding a protein homologous to human NIP3. Genes Chromosomes Cancer 1998, 21, 230–235. [Google Scholar] [CrossRef]
- Sandoval, H.; Thiagarajan, P.; Dasgupta, S.K.; Schumacher, A.; Prchal, J.T.; Chen, M.; Wang, J. Essential role for Nix in autophagic maturation of erythroid cells. Nature 2008, 454, 232–235. [Google Scholar] [CrossRef]
- Schweers, R.L.; Zhang, J.; Randall, M.S.; Loyd, M.R.; Li, W.; Dorsey, F.C.; Kundu, M.; Opferman, J.T.; Cleveland, J.L.; Miller, J.L.; et al. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc. Natl. Acad. Sci. USA 2007, 104, 19500–19505. [Google Scholar] [CrossRef] [Green Version]
- O’Sullivan, T.E.; Johnson, L.R.; Kang, H.H.; Sun, J.C. BNIP3- and BNIP3L-Mediated Mitophagy Promotes the Generation of Natural Killer Cell Memory. Immunity 2015, 43, 331–342. [Google Scholar] [CrossRef] [Green Version]
- Esteban-Martinez, L.; Boya, P. BNIP3L/NIX-dependent mitophagy regulates cell differentiation via metabolic reprogramming. Autophagy 2018, 14, 915–917. [Google Scholar] [CrossRef]
- Liu, L.; Feng, D.; Chen, G.; Chen, M.; Zheng, Q.; Song, P.; Ma, Q.; Zhu, C.; Wang, R.; Qi, W.; et al. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat. Cell Biol. 2012, 14, 177–185. [Google Scholar] [CrossRef]
- Wu, H.; Xue, D.; Chen, G.; Han, Z.; Huang, L.; Zhu, C.; Wang, X.; Jin, H.; Wang, J.; Zhu, Y.; et al. The BCL2L1 and PGAM5 axis defines hypoxia-induced receptor-mediated mitophagy. Autophagy 2014, 10, 1712–1725. [Google Scholar] [CrossRef] [Green Version]
- Wu, W.; Tian, W.; Hu, Z.; Chen, G.; Huang, L.; Li, W.; Zhang, X.; Xue, P.; Zhou, C.; Liu, L.; et al. ULK1 translocates to mitochondria and phosphorylates FUNDC1 to regulate mitophagy. EMBO Rep. 2014, 15, 566–575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, G.; Han, Z.; Feng, D.; Chen, Y.; Chen, L.; Wu, H.; Huang, L.; Zhou, C.; Cai, X.; Fu, C.; et al. A regulatory signaling loop comprising the PGAM5 phosphatase and CK2 controls receptor-mediated mitophagy. Mol. Cell 2014, 54, 362–377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miranda-Diaz, A.G.; Cardona-Munoz, E.G.; Pacheco-Moises, F.P. The Role of Cardiolipin and Mitochondrial Damage in Kidney Transplant. Oxid. Med. Cell. Longev. 2019, 2019, 3836186. [Google Scholar] [CrossRef]
- Paradies, G.; Paradies, V.; Ruggiero, F.M.; Petrosillo, G. Role of Cardiolipin in Mitochondrial Function and Dynamics in Health and Disease: Molecular and Pharmacological Aspects. Cells 2019, 8, 728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tschopp, J.; Schroder, K. NLRP3 inflammasome activation: The convergence of multiple signalling pathways on ROS production? Nat. Rev. Immunol. 2010, 10, 210–215. [Google Scholar] [CrossRef] [PubMed]
- Anton, Z.; Landajuela, A.; Hervas, J.H.; Montes, L.R.; Hernandez-Tiedra, S.; Velasco, G.; Goni, F.M.; Alonso, A. Human Atg8-cardiolipin interactions in mitophagy: Specific properties of LC3B, GABARAPL2 and GABARAP. Autophagy 2016, 12, 2386–2403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chu, C.T.; Bayir, H.; Kagan, V.E. LC3 binds externalized cardiolipin on injured mitochondria to signal mitophagy in neurons: Implications for Parkinson disease. Autophagy 2014, 10, 376–378. [Google Scholar] [CrossRef] [Green Version]
- Chu, C.T.; Ji, J.; Dagda, R.K.; Jiang, J.F.; Tyurina, Y.Y.; Kapralov, A.A.; Tyurin, V.A.; Yanamala, N.; Shrivastava, I.H.; Mohammadyani, D.; et al. Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nat. Cell. Biol. 2013, 15, 1197–1205. [Google Scholar] [CrossRef] [Green Version]
- Shen, Z.; Li, Y.; Gasparski, A.N.; Abeliovich, H.; Greenberg, M.L. Cardiolipin Regulates Mitophagy through the Protein Kinase C Pathway. J. Biol. Chem. 2017, 292, 2916–2923. [Google Scholar] [CrossRef] [Green Version]
- Hsu, P.; Liu, X.; Zhang, J.; Wang, H.G.; Ye, J.M.; Shi, Y. Cardiolipin remodeling by TAZ/tafazzin is selectively required for the initiation of mitophagy. Autophagy 2015, 11, 643–652. [Google Scholar] [CrossRef] [Green Version]
- Sun, Q.; Fan, J.; Billiar, T.R.; Scott, M.J. Inflammasome and autophagy regulation - a two-way street. Mol. Med. 2017, 23, 188–195. [Google Scholar] [CrossRef] [PubMed]
- Marcuzzi, A.; Piscianz, E.; Loganes, C.; Vecchi Brumatti, L.; Knowles, A.; Bilel, S.; Tommasini, A.; Bortul, R.; Zweyer, M. Innovative Target Therapies Are Able to Block the Inflammation Associated with Dysfunction of the Cholesterol Biosynthesis Pathway. Int. J. Mol. Sci. 2015, 17, 47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, R.; Yazdi, A.S.; Menu, P.; Tschopp, J. A role for mitochondria in NLRP3 inflammasome activation. Nature 2011, 469, 221–225. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.S.; Suh, H.W.; Kim, S.J.; Jo, E.K. Mitochondrial Control of Innate Immunity and Inflammation. Immune Netw. 2017, 17, 77–88. [Google Scholar] [CrossRef] [Green Version]
- Helal, J.N.; Van Hoecke, J.; Garapon-Bar, C.; Goubel, F. Surface myoelectric signals during ergocycle exrcises at various mechanical powers and pedalling rates. J. Electromyogr. Kinesiol. 1992, 2, 242–251. [Google Scholar] [CrossRef]
- Tan, T.; Zimmermann, M.; Reichert, A.S. Controlling quality and amount of mitochondria by mitophagy: Insights into the role of ubiquitination and deubiquitination. Biol. Chem. 2016, 397, 637–647. [Google Scholar] [CrossRef] [PubMed]
- Picca, A.; Lezza, A.M.S.; Leeuwenburgh, C.; Pesce, V.; Calvani, R.; Landi, F.; Bernabei, R.; Marzetti, E. Fueling Inflamm-Aging through Mitochondrial Dysfunction: Mechanisms and Molecular Targets. Int. J. Mol. Sci. 2017, 18, 933. [Google Scholar] [CrossRef]
- Xu, Y.; Shen, J.; Ran, Z. Emerging views of mitophagy in immunity and autoimmune diseases. Autophagy 2020, 16, 3–17. [Google Scholar] [CrossRef]
- Rottenberg, H.; Hoek, J.B. The path from mitochondrial ROS to aging runs through the mitochondrial permeability transition pore. Aging Cell 2017, 16, 943–955. [Google Scholar] [CrossRef] [Green Version]
- Harrington, J.S.; Choi, A.M.K.; Nakahira, K. Mitochondrial DNA in Sepsis. Curr. Opin. Crit. Care 2017, 23, 284–290. [Google Scholar] [CrossRef]
- Dudek, J. Role of Cardiolipin in Mitochondrial Signaling Pathways. Front. Cell. Dev. Biol. 2017, 5, 90. [Google Scholar] [CrossRef] [Green Version]
- Schlattner, U.; Tokarska-Schlattner, M.; Ramirez, S.; Tyurina, Y.Y.; Amoscato, A.A.; Mohammadyani, D.; Huang, Z.; Jiang, J.; Yanamala, N.; Seffouh, A.; et al. Dual function of mitochondrial Nm23-H4 protein in phosphotransfer and intermembrane lipid transfer: A cardiolipin-dependent switch. J. Biol. Chem. 2013, 288, 111–121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lacombe, M.L.; Tokarska-Schlattner, M.; Epand, R.F.; Boissan, M.; Epand, R.M.; Schlattner, U. Interaction of NDPK-D with cardiolipin-containing membranes: Structural basis and implications for mitochondrial physiology. Biochimie 2009, 91, 779–783. [Google Scholar] [CrossRef]
- Zhong, Z.; Umemura, A.; Sanchez-Lopez, E.; Liang, S.; Shalapour, S.; Wong, J.; He, F.; Boassa, D.; Perkins, G.; Ali, S.R.; et al. NF-kappaB Restricts Inflammasome Activation via Elimination of Damaged Mitochondria. Cell 2016, 164, 896–910. [Google Scholar] [CrossRef] [Green Version]
- Yeon, S.H.; Yang, G.; Lee, H.E.; Lee, J.Y. Oxidized phosphatidylcholine induces the activation of NLRP3 inflammasome in macrophages. J. Leukoc. Biol. 2017, 101, 205–215. [Google Scholar] [CrossRef]
- Zhang, Q.; Kuang, H.; Chen, C.; Yan, J.; Do-Umehara, H.C.; Liu, X.Y.; Dada, L.; Ridge, K.M.; Chandel, N.S.; Liu, J. The kinase Jnk2 promotes stress-induced mitophagy by targeting the small mitochondrial form of the tumor suppressor ARF for degradation. Nat. Immunol. 2015, 16, 458–466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Xu, X.; Ma, J.; Wu, J.; Wang, Y.; Zhou, R.; Han, J. Gene deletion of Gabarap enhances Nlrp3 inflammasome-dependent inflammatory responses. J. Immunol. 2013, 190, 3517–3524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanchez-Lopez, E.; Zhong, Z.; Stubelius, A.; Sweeney, S.R.; Booshehri, L.M.; Antonucci, L.; Liu-Bryan, R.; Lodi, A.; Terkeltaub, R.; Lacal, J.C.; et al. Choline Uptake and Metabolism Modulate Macrophage IL-1beta and IL-18 Production. Cell Metab. 2019, 29, 1350–1362 e1357. [Google Scholar] [CrossRef]
- Li, S.; Wu, H.; Han, D.; Ma, S.; Fan, W.; Wang, Y.; Zhang, R.; Fan, M.; Huang, Y.; Fu, X.; et al. A Novel Mechanism of Mesenchymal Stromal Cell-Mediated Protection against Sepsis: Restricting Inflammasome Activation in Macrophages by Increasing Mitophagy and Decreasing Mitochondrial ROS. Oxid. Med. Cell. Longev. 2018, 2018, 3537609. [Google Scholar] [CrossRef]
- Cho, D.H.; Kim, J.K.; Jo, E.K. Mitophagy and Innate Immunity in Infection. Mol. Cells 2020, 43, 10–22. [Google Scholar] [CrossRef]
- Jabir, M.S.; Hopkins, L.; Ritchie, N.D.; Ullah, I.; Bayes, H.K.; Li, D.; Tourlomousis, P.; Lupton, A.; Puleston, D.; Simon, A.K.; et al. Mitochondrial damage contributes to Pseudomonas aeruginosa activation of the inflammasome and is downregulated by autophagy. Autophagy 2015, 11, 166–182. [Google Scholar] [CrossRef] [Green Version]
- Li, R.; Fang, L.; Tan, S.; Yu, M.; Li, X.; He, S.; Wei, Y.; Li, G.; Jiang, J.; Wu, M. Type I CRISPR-Cas targets endogenous genes and regulates virulence to evade mammalian host immunity. Cell Res. 2016, 26, 1273–1287. [Google Scholar] [CrossRef]
- Wu, Q.; Wang, B.; Zhou, C.; Lin, P.; Qin, S.; Gao, P.; Wang, Z.; Xia, Z.; Wu, M. Bacterial Type I CRISPR-Cas systems influence inflammasome activation in mammalian host by promoting autophagy. Immunology 2019, 158, 240–251. [Google Scholar] [CrossRef]
- Kader, M.; Alaoui-El-Azher, M.; Vorhauer, J.; Kode, B.B.; Wells, J.Z.; Stolz, D.; Michalopoulos, G.; Wells, A.; Scott, M.; Ismail, N. MyD88-dependent inflammasome activation and autophagy inhibition contributes to Ehrlichia-induced liver injury and toxic shock. PLoS Pathog. 2017, 13, e1006644. [Google Scholar] [CrossRef] [Green Version]
- Xia, B.; Yu, J.; He, T.; Liu, X.; Su, J.; Wang, M.; Wang, J.; Zhu, Y. Lactobacillus johnsonii L531 ameliorates enteritis via elimination of damaged mitochondria and suppression of SQSTM1-dependent mitophagy in a Salmonella infantis model of piglet diarrhea. FASEB J. 2020, 34, 2821–2839. [Google Scholar] [CrossRef] [Green Version]
- Lupfer, C.; Thomas, P.G.; Anand, P.K.; Vogel, P.; Milasta, S.; Martinez, J.; Huang, G.; Green, M.; Kundu, M.; Chi, H.; et al. Receptor interacting protein kinase 2-mediated mitophagy regulates inflammasome activation during virus infection. Nat. Immunol. 2013, 14, 480–488. [Google Scholar] [CrossRef] [Green Version]
- Ojeda, D.S.; Grasso, D.; Urquiza, J.; Till, A.; Vaccaro, M.I.; Quarleri, J. Cell Death Is Counteracted by Mitophagy in HIV-Productively Infected Astrocytes but Is Promoted by Inflammasome Activation Among Non-productively Infected Cells. Front. Immunol. 2018, 9, 2633. [Google Scholar] [CrossRef] [Green Version]
- Rawat, P.; Teodorof-Diedrich, C.; Spector, S.A. Human immunodeficiency virus Type-1 single-stranded RNA activates the NLRP3 inflammasome and impairs autophagic clearance of damaged mitochondria in human microglia. Glia 2019, 67, 802–824. [Google Scholar] [CrossRef] [PubMed]
- Thangaraj, A.; Periyasamy, P.; Liao, K.; Bendi, V.S.; Callen, S.; Pendyala, G.; Buch, S. HIV-1 TAT-mediated microglial activation: Role of mitochondrial dysfunction and defective mitophagy. Autophagy 2018, 14, 1596–1619. [Google Scholar] [CrossRef] [Green Version]
- Chivero, E.T.; Guo, M.L.; Periyasamy, P.; Liao, K.; Callen, S.E.; Buch, S. HIV-1 Tat Primes and Activates Microglial NLRP3 Inflammasome-Mediated Neuroinflammation. J. Neurosci. 2017, 37, 3599–3609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mamik, M.K.; Hui, E.; Branton, W.G.; McKenzie, B.A.; Chisholm, J.; Cohen, E.A.; Power, C. HIV-1 Viral Protein R Activates NLRP3 Inflammasome in Microglia: Implications for HIV-1 Associated Neuroinflammation. J. Neuroimmune Pharmacol. 2017, 12, 233–248. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Ma, C.; Long, F.; Yang, D.; Liu, X.; Hu, Y.; Wu, C.; Wang, B.; Wang, M.; Chen, Y.; et al. Parkin Impairs Antiviral Immunity by Suppressing the Mitochondrial Reactive Oxygen Species-Nlrp3 Axis and Antiviral Inflammation. iScience 2019, 16, 468–484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, S.; Xia, C.; Li, S.; Du, L.; Zhang, L.; Zhou, R. Defective mitophagy driven by dysregulation of rheb and KIF5B contributes to mitochondrial reactive oxygen species (ROS)-induced nod-like receptor 3 (NLRP3) dependent proinflammatory response and aggravates lipotoxicity. Redox Biol. 2014, 3, 63–71. [Google Scholar] [CrossRef] [Green Version]
- Durga Devi, T.; Babu, M.; Makinen, P.; Kaikkonen, M.U.; Heinaniemi, M.; Laakso, H.; Yla-Herttuala, E.; Rieppo, L.; Liimatainen, T.; Naumenko, N.; et al. Aggravated Postinfarct Heart Failure in Type 2 Diabetes Is Associated with Impaired Mitophagy and Exaggerated Inflammasome Activation. Am. J. Pathol. 2017, 187, 2659–2673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiu, B.; Jantuan, E.; Shen, F.; Chiu, B.; Sergi, C. Autophagy-Inflammasome Interplay in Heart Failure: A Systematic Review on Basics, Pathways, and Therapeutic Perspectives. Ann. Clin. Lab. Sci. 2017, 47, 243–252. [Google Scholar]
- Ghosh, R.; Pattison, J.S. Macroautophagy and Chaperone-Mediated Autophagy in Heart Failure: The Known and the Unknown. Oxidative Med. Cell. Longev. 2018, 2018, 8602041. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Miao, W.; Ma, J.; Xv, Z.; Bo, H.; Li, J.; Zhang, Y.; Ji, L.L. Acute Exercise-Induced Mitochondrial Stress Triggers an Inflammatory Response in the Myocardium via NLRP3 Inflammasome Activation with Mitophagy. Oxidative Med. Cell. Longev. 2016, 2016, 1987149. [Google Scholar] [CrossRef] [Green Version]
- Onat, U.I.; Yildirim, A.D.; Tufanli, O.; Cimen, I.; Kocaturk, B.; Veli, Z.; Hamid, S.M.; Shimada, K.; Chen, S.; Sin, J.; et al. Intercepting the Lipid-Induced Integrated Stress Response Reduces Atherosclerosis. J. Am. Coll. Cardiol. 2019, 73, 1149–1169. [Google Scholar] [CrossRef]
- Zhang, N.P.; Liu, X.J.; Xie, L.; Shen, X.Z.; Wu, J. Impaired mitophagy triggers NLRP3 inflammasome activation during the progression from nonalcoholic fatty liver to nonalcoholic steatohepatitis. Lab. Invest. 2019, 99, 749–763. [Google Scholar] [CrossRef]
- Gao, Y.; Li, J.; Li, J.; Hu, C.; Zhang, L.; Yan, J.; Li, L.; Zhang, L. Tetrahydroxy stilbene glycoside alleviated inflammatory damage by mitophagy via AMPK related PINK1/Parkin signaling pathway. Biochem. Pharmacol. 2020, 177, 113997. [Google Scholar] [CrossRef]
- Hu, Z.L.; Sun, T.; Lu, M.; Ding, J.H.; Du, R.H.; Hu, G. Kir6.1/K-ATP channel on astrocytes protects against dopaminergic neurodegeneration in the MPTP mouse model of Parkinson’s disease via promoting mitophagy. Brain. Behav. Immun. 2019, 81, 509–522. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Wise, L.; Fukuchi, K.I. TLR4 Cross-Talk With NLRP3 Inflammasome and Complement Signaling Pathways in Alzheimer’s Disease. Front. Immunol. 2020, 11, 724. [Google Scholar] [CrossRef] [PubMed]
- Su, S.H.; Wu, Y.F.; Lin, Q.; Wang, D.P.; Hai, J. URB597 protects against NLRP3 inflammasome activation by inhibiting autophagy dysfunction in a rat model of chronic cerebral hypoperfusion. J. Neuroinflammation 2019, 16, 260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tseng, W.A.; Thein, T.; Kinnunen, K.; Lashkari, K.; Gregory, M.S.; D’Amore, P.A.; Ksander, B.R. NLRP3 inflammasome activation in retinal pigment epithelial cells by lysosomal destabilization: Implications for age-related macular degeneration. Invest. Ophthalmol. Vis. Sci. 2013, 54, 110–120. [Google Scholar] [CrossRef]
- Lin, Q.; Li, S.; Jiang, N.; Shao, X.; Zhang, M.; Jin, H.; Zhang, Z.; Shen, J.; Zhou, Y.; Zhou, W.; et al. PINK1-parkin pathway of mitophagy protects against contrast-induced acute kidney injury via decreasing mitochondrial ROS and NLRP3 inflammasome activation. Redox Biol. 2019, 26, 101254. [Google Scholar] [CrossRef]
- Xu, Y.; Wang, J.; Xu, W.; Ding, F.; Ding, W. Prohibitin 2-mediated mitophagy attenuates renal tubular epithelial cells injury by regulating mitochondrial dysfunction and NLRP3 inflammasome activation. Am. J. Physiol. Renal Physiol. 2019, 316, F396–F407. [Google Scholar] [CrossRef]
- Kang, R.; Zeng, L.; Xie, Y.; Yan, Z.; Zhou, B.; Cao, L.; Klionsky, D.J.; Tracey, K.J.; Li, J.; Wang, H.; et al. A novel PINK1- and PARK2-dependent protective neuroimmune pathway in lethal sepsis. Autophagy 2016, 12, 2374–2385. [Google Scholar] [CrossRef] [Green Version]
- Drake, L.E.; Springer, M.Z.; Poole, L.P.; Kim, C.J.; Macleod, K.F. Expanding perspectives on the significance of mitophagy in cancer. Semin. Cancer Biol. 2017, 47, 110–124. [Google Scholar] [CrossRef]
- Li, C.; Zhang, Y.; Cheng, X.; Yuan, H.; Zhu, S.; Liu, J.; Wen, Q.; Xie, Y.; Liu, J.; Kroemer, G.; et al. PINK1 and PARK2 Suppress Pancreatic Tumorigenesis through Control of Mitochondrial Iron-Mediated Immunometabolism. Dev. Cell 2018, 46, 441–455 e448. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Li, Y.; Siraj, S.; Jin, H.; Fan, Y.; Yang, X.; Huang, X.; Wang, X.; Wang, J.; Liu, L.; et al. FUN14 Domain-Containing 1-Mediated Mitophagy Suppresses Hepatocarcinogenesis by Inhibition of Inflammasome Activation in Mice. Hepatology 2019, 69, 604–621. [Google Scholar] [CrossRef]
- Bhansali, S.; Bhansali, A.; Dutta, P.; Walia, R.; Dhawan, V. Metformin upregulates mitophagy in patients with T2DM: A randomized placebo-controlled study. J. Cell. Mol. Med. 2020, 24, 2832–2846. [Google Scholar] [CrossRef]
- Iacano, A.J.; Lewis, H.; Hazen, J.E.; Andro, H.; Smith, J.D.; Gulshan, K. Miltefosine increases macrophage cholesterol release and inhibits NLRP3-inflammasome assembly and IL-1beta release. Sci. Rep. 2019, 9, 11128. [Google Scholar] [CrossRef] [Green Version]
- Ma, S.; Chen, J.; Feng, J.; Zhang, R.; Fan, M.; Han, D.; Li, X.; Li, C.; Ren, J.; Wang, Y.; et al. Melatonin Ameliorates the Progression of Atherosclerosis via Mitophagy Activation and NLRP3 Inflammasome Inhibition. Oxidative Med. Cell. Longev. 2018, 2018, 9286458. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.; Shrestha, S.; Li, J.; Yu, X.; Chen, J.; Yan, F.; Ying, G.; Gu, C.; Wang, L.; Chen, G. Melatonin-mediated mitophagy protects against early brain injury after subarachnoid hemorrhage through inhibition of NLRP3 inflammasome activation. Sci. Rep. 2017, 7, 2417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, X.; Hao, M.; Liu, Y.; Ma, X.; Lin, W.; Xu, Q.; Zhou, H.; Shao, N.; Kuang, H. Liraglutide ameliorates non-alcoholic steatohepatitis by inhibiting NLRP3 inflammasome and pyroptosis activation via mitophagy. Eur. J. Pharmacol. 2019, 864, 172715. [Google Scholar] [CrossRef] [PubMed]
- Szeto, H.H.; Liu, S.; Soong, Y.; Seshan, S.V.; Cohen-Gould, L.; Manichev, V.; Feldman, L.C.; Gustafsson, T. Mitochondria Protection after Acute Ischemia Prevents Prolonged Upregulation of IL-1beta and IL-18 and Arrests CKD. J. Am. Soc. Nephrol. 2017, 28, 1437–1449. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.J.; Bae, S.H.; Ryu, J.C.; Kwon, Y.; Oh, J.H.; Kwon, J.; Moon, J.S.; Kim, K.; Miyawaki, A.; Lee, M.G.; et al. SESN2/sestrin2 suppresses sepsis by inducing mitophagy and inhibiting NLRP3 activation in macrophages. Autophagy 2016, 12, 1272–1291. [Google Scholar] [CrossRef] [Green Version]
- Mai, C.T.; Wu, M.M.; Wang, C.L.; Su, Z.R.; Cheng, Y.Y.; Zhang, X.J. Palmatine attenuated dextran sulfate sodium (DSS)-induced colitis via promoting mitophagy-mediated NLRP3 inflammasome inactivation. Mol Immunol. 2019, 105, 76–85. [Google Scholar] [CrossRef]
- Liu, C.; Wang, J.; Yang, Y.; Liu, X.; Zhu, Y.; Zou, J.; Peng, S.; Le, T.H.; Chen, Y.; Zhao, S.; et al. Ginsenoside Rd ameliorates colitis by inducing p62-driven mitophagy-mediated NLRP3 inflammasome inactivation in mice. Biochem. Pharmacol. 2018, 155, 366–379. [Google Scholar] [CrossRef]
- Guo, W.; Sun, Y.; Liu, W.; Wu, X.; Guo, L.; Cai, P.; Wu, X.; Wu, X.; Shen, Y.; Shu, Y.; et al. Small molecule-driven mitophagy-mediated NLRP3 inflammasome inhibition is responsible for the prevention of colitis-associated cancer. Autophagy 2014, 10, 972–985. [Google Scholar] [CrossRef]
- Wu, J.; Li, X.; Zhu, G.; Zhang, Y.; He, M.; Zhang, J. The role of Resveratrol-induced mitophagy/autophagy in peritoneal mesothelial cells inflammatory injury via NLRP3 inflammasome activation triggered by mitochondrial ROS. Exp. Cell Res. 2016, 341, 42–53. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.S.; Chen, L.; Lu, Y.Y.; Lei, S.Q.; Peng, M.; Xia, Z.Y. Honokiol-Mediated Mitophagy Ameliorates Postoperative Cognitive Impairment Induced by Surgery/Sevoflurane via Inhibiting the Activation of NLRP3 Inflammasome in the Hippocampus. Oxidative Med. Cell. Longev. 2019, 2019, 8639618. [Google Scholar] [CrossRef] [PubMed]
- Su, S.H.; Wang, Y.Q.; Wu, Y.F.; Wang, D.P.; Lin, Q.; Hai, J. Cannabinoid receptor agonist WIN55,212-2 and fatty acid amide hydrolase inhibitor URB597 may protect against cognitive impairment in rats of chronic cerebral hypoperfusion via PI3K/AKT signaling. Behav. Brain Res. 2016, 313, 334–344. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Meng, J.; Xu, Q.; Long, T.; Bi, F.; Chang, C.; Liu, W. Rapamycin improves the neuroprotection effect of inhibition of NLRP3 inflammasome activation after TBI. Brain Res. 2019, 1710, 163–172. [Google Scholar] [CrossRef]
- He, Q.; Li, Z.; Meng, C.; Wu, J.; Zhao, Y.; Zhao, J. Parkin-Dependent Mitophagy is Required for the Inhibition of ATF4 on NLRP3 Inflammasome Activation in Cerebral Ischemia-Reperfusion Injury in Rats. Cells 2019, 8, 897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yumnamcha, T.; Devi, T.S.; Singh, L.P. Auranofin Mediates Mitochondrial Dysregulation and Inflammatory Cell Death in Human Retinal Pigment Epithelial Cells: Implications of Retinal Neurodegenerative Diseases. Front. Neurosci. 2019, 13, 1065. [Google Scholar] [CrossRef]
- Pillai, V.B.; Samant, S.; Sundaresan, N.R.; Raghuraman, H.; Kim, G.; Bonner, M.Y.; Arbiser, J.L.; Walker, D.I.; Jones, D.P.; Gius, D.; et al. Honokiol blocks and reverses cardiac hypertrophy in mice by activating mitochondrial Sirt3. Nat. Commun. 2015, 6, 6656. [Google Scholar] [CrossRef]
- Woldt, E.; Sebti, Y.; Solt, L.A.; Duhem, C.; Lancel, S.; Eeckhoute, J.; Hesselink, M.K.; Paquet, C.; Delhaye, S.; Shin, Y.; et al. Rev-erb-alpha modulates skeletal muscle oxidative capacity by regulating mitochondrial biogenesis and autophagy. Nat. Med. 2013, 19, 1039–1046. [Google Scholar] [CrossRef]
- Wang, S.; Lin, Y.; Yuan, X.; Li, F.; Guo, L.; Wu, B. REV-ERBalpha integrates colon clock with experimental colitis through regulation of NF-kappaB/NLRP3 axis. Nat. Commun. 2018, 9, 4246. [Google Scholar] [CrossRef] [Green Version]
- Pourcet, B.; Zecchin, M.; Ferri, L.; Beauchamp, J.; Sitaula, S.; Billon, C.; Delhaye, S.; Vanhoutte, J.; Mayeuf-Louchart, A.; Thorel, Q.; et al. Nuclear Receptor Subfamily 1 Group D Member 1 Regulates Circadian Activity of NLRP3 Inflammasome to Reduce the Severity of Fulminant Hepatitis in Mice. Gastroenterology 2018, 154, 1449–1464. [Google Scholar] [CrossRef]
- Zhou, Z.; Lin, Y.; Gao, L.; Yang, Z.; Wang, S.; Wu, B. Circadian pharmacological effects of berberine on chronic colitis in mice: Role of the clock component Rev-erbalpha. Biochem. Pharmacol. 2020, 172, 113773. [Google Scholar] [CrossRef] [PubMed]
- Reitz, C.J.; Alibhai, F.J.; Khatua, T.N.; Rasouli, M.; Bridle, B.W.; Burris, T.P.; Martino, T.A. SR9009 administered for one day after myocardial ischemia-reperfusion prevents heart failure in mice by targeting the cardiac inflammasome. Commun. Biol. 2019, 2, 353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, D.; Fang, X.; Xu, Y.; Xiao, H.; Huang, T.; Zhang, Y.; Ge, Y.; Li, Y.; Zong, L.; Gao, J. Rev-erbalpha can regulate the NF-kappaB/NALP3 pathway to modulate lipopolysaccharide-induced acute lung injury and inflammation. Int. Immunopharmacol. 2019, 73, 312–320. [Google Scholar] [CrossRef] [PubMed]



| Agents/Drugs | Disease/Model | Functions | Underlying Mechanisms | Experimental Objects | Reference |
|---|---|---|---|---|---|
| Metformin | Type 2 diabetes | ↓ Mitochondrial oxidative stress and distortions in mitochondrial morphology | ↑ Protein expression of p-AMPKα (T172), and NLRP3 ↑ Protein expression of mitophagy-related markers (PINK1, PARKIN, MFN2, NIX, LC3-II, LAMP2) | Patient samples, PBMCs | [171] |
| Miltefosine | Atherosclerosis | ↑ ABCA1-mediated cholesterol release ↓ LPS-mediated NLRP3 inflammasome assembly and IL-1β release ↑ Mitophagy induction | ↑ Disruption of lipid-rafts ↑ Inhibition of phosphatidylserine flip ↓ LPS-mediated mitochondrial ROS generation ↓ Loss of mitochondrial membrane potential in murine macrophages treated with LPS | HEK293-ABCA1-GFP cells, BHK cells, RAW264.7, BMDM | [172] |
| Melatonin | Atherosclerosis | ↓ Atherosclerotic plaque progression and serum IL-1β level in vivo murine models ↓ NLRP3 inflammasome activation in atherosclerotic lesions and in Ox-LDL-treated murine macrophages | ↑ Sirt3-dependent mitophagy induction and reactive oxygen species (ROS) scavenging ↑ Parkin-mediated mitophagy process through Sirt3-FOXO3a pathway | RAW264.7, ApoE−/− mice | [173] |
| Subarachnoid hemorrhage | ↓ Brain edema and neurological dysfunction in rat receiving SAH ↓ SAH-induced neuronal cell death in the ipsilateral basal cortex ↓ Pathological changes in mitochondria | ↑ Protein expression of autophagy markers (LC3-II/LC3-I and Atg5) and mitophagy markers (Parkin and PINK-1) ↓ ROS and pro-inflammatory cytokines generation ↓ NLRP3 inflammasome activation | Endovascular perforation-induced SAH rat model, Brain tissue | [174] | |
| Liraglutide | Non-alcoholic steatohepatitis | ↓ Lipid accumulation in hepatocytes ↓ NLRP3 inflammasome-induced hepatocyte pyroptosis ↑ Mitophagy induction | ↓ Mitochondrial dysfunction and reactive oxygen species generation ↑ Co-localization of PINK1/Parkin and mitochondria and OPTN and NIX expressions in HepG2 cells treated with LPS and PA | HepG2 cell line | [175] |
| URB597 | Chronic cerebral hypoperfusion | ↓ CCH-induced NLRP3 inflammasome activation, impaired autophagy, and defective mitophagy in rat hippocampus | ↑ The restoration of lysosomal function ↓ CCH-induced microglial overactivation ↓ CCH-induced ROS accumulation | BCCAo-induced CCH rat model, Hippocampus | [163] |
| Elamipretide (SS-31) | Bilateral renal ischemia | ↓ Ischemia-induced tubulointerstitial fibrosis and glomerulosclerosis ↓ Inflammatory Response and endothelial Injury after Ischemia | ↓ NLRP3 inflammasome activation and upregulation of IL-18, IL-1β, and TNF induced by ischemia ↑ Mitochondrial integrity and autophagic vacuoles containing organelles and cytoplasmic contents in podocytes | Ischemia-reperfusion experimental rat model, Kidney tissue | [176] |
| SESN2 | Sepsis | ↓ Clearance of damaged mitochondria in sesn2−/− macrophages ↑ IL1β and IL18 production in serum and lung tissue of sesn2−/− mice after CLP ↑ Mortality rate in sesn2−/− mice | ↑ Mitochondrial priming by mediating aggregation of SQSTM1 and its binding to lysine 63-linked ubiquitins on the mitochondrial surface ↑ Activation of autophagic machinery for the degradation of primed mitochondria via an increase of ULK1 protein ↓ Prolonged NLRP3 inflammasome activation by inducing mitophagy | sesn2−/− mice, sesn2−/− GFPM -AP1-LC3 mice, nos2−/− mice, nlrp3−/− mice, BMDM | [177] |
| Palmatine | Ulcerative colitis | ↓ Pathological and histological scores in DSS mice · ↓ Colonic inflammation in DSS mice | ↓ NLRP3 inflammasomes activation in DSS mice and THP-1 ↑ PINK1 and Parkin-driven mitophagy in DSS mice and THP-1 | DSS-induced UC model using C57BL/6 mice, THP-1 cell | [178] |
| Ginsenoside Rd | Ulcerative colitis | ↓ Body weight loss, stool consistency alterations and blood loss in DSS mice ↓ Recruitment of inflammatory cell into colonic tissue ↓ NLRP3 inflammasome activation in colon tissue and THP-1 cells | ↓ LPS plus ATP-induced mitochondrial perturbation and mtDNA release in THP-1 ↑ p62 expression and its translocation to mitochondria ↑ p62-mediated autophagy activation ↑ AMPK/ULK1signaling pathway in vitro and in vivo | DSS-induced UC model using BALB/C mice, THP-1 cell, Colonic cell suspension | [179] |
| Pramipexole | Sepsis | ↑ Survival rate in septic pink1-/- and park2-/- mice ↓ Inflammasome-dependent cytokines (IL-1β and HMGB1) in septic pink1-/- and park2-/- mice | ↑ Circulating dopamine levels in septic pink1-/- and park2-/- mice ↓ Circulating lactate levels in septic pink1-/- and park2-/- mice ↓ Sepsis-induced Hif1α, Ldha, and Pdk1 as well as GSH depletion in the brain from pink1-/- and park2-/- mice ↓ Hif1α-mediated aerobic glycolysis involved in inflammasome activation in pink1-/- and park2-/- mice | CLP sepsis in pink1-/-, park2-/-, nlrp3-/-, hif1α-/- mice | [167] |
| Andrographolide | Colitis-associated cancer | ↓ Tumorigenesis and inflammation in a colitis-associated colorectal cancer model ↓ Progression of DSS-induced experimental colitis in mice | ↓ NLRP3 inflammasome activation in LPS-primed macrophages treated with ATP or in mice with DSS-induced colitis ↓ ATP-induced collapse of the mitochondria membrane potential and mitochondria fragmentation in LPS-primed macrophages ↑ Mitophagy induction in macrophages | THP-1 cell, Peritoneal macrophages, BMDM, In vivo CAC mice models | [180] |
| Resveratrol | Peritoneal mesothelial cells inflammatory injury | ↑ Protection from ROS-NLRP3-mediated inflammatory injury | ↑ Mitophagy/autophagy via AMPK activation | Human peritoneal mesothelial cell line (HMrSV5) | [181] |
| Honokiol | Behavioral and cognitive impairment | ↑ Cognitive recovery | ↑ Mitophagy ↓ mtROS NLRP3 and microglia activation in the hippocampus ↓ Neuronal apoptosis | Surgery/anesthesia (sevoflurane) induced cognitive impairment in mice | [182] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuk, J.-M.; Silwal, P.; Jo, E.-K. Inflammasome and Mitophagy Connection in Health and Disease. Int. J. Mol. Sci. 2020, 21, 4714. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21134714
Yuk J-M, Silwal P, Jo E-K. Inflammasome and Mitophagy Connection in Health and Disease. International Journal of Molecular Sciences. 2020; 21(13):4714. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21134714
Chicago/Turabian StyleYuk, Jae-Min, Prashanta Silwal, and Eun-Kyeong Jo. 2020. "Inflammasome and Mitophagy Connection in Health and Disease" International Journal of Molecular Sciences 21, no. 13: 4714. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21134714