The Arabidopsis RLCK VI_A2 Kinase Controls Seedling and Plant Growth in Parallel with Gibberellin
Abstract
:1. Introduction
2. Results
2.1. Molecular Characterization of the RLCK VI_A2 T-DNA Insertion Mutant and the Transgenic Plants Used in the Study
2.2. The RLCK VI_A2 Kinase Controls Seedling and Plant Growth
2.3. The RLCK VI_A2 Kinase Controls Cell Size
2.4. Gibberellic Acid Treatment Rectifies the rlck vi_a2 Mutant Phenotypes
2.5. The Effect of the rlck vi_a2 Mutation on Gibberellic Acid Level, Synthesis, and Signalling in Seedlings
2.6. Transcriptome Analysis Indicated a Gibberellin-Independent Role for the RLCK VI_A2 Kinase in Skotomorphogenesis
2.7. Transcriptome Analysis Revealed the Role of the RLCK VI_A2 Kinase in Cellular Transport and Cell Wall Organisation
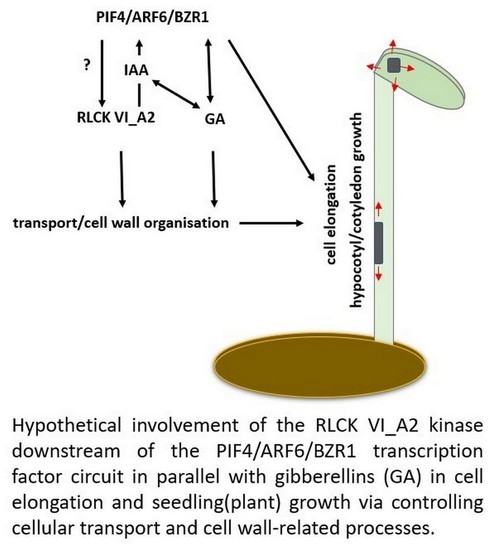
3. Discussion
3.1. The RLCK VI_A Kinases Are Required for Cell Elongation and Organ Growth in Addition to Their Role in Stress Responses
3.2. How the RLCK VI_A2 Kinase May Affect Cell Expansion?
3.3. Gibberellin might Indirectly Complement for the Missing Kinase Function
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Analysis of Hypocotyl Length and Rosette Size Measurement
4.3. Cell Size and Shape Analysis
4.4. Gibberellin Content Measurement by Ultra-High Performance Liquid Chromatography-Tandem Mass Spectrometry
4.5. Hormone Treatments
4.6. Characterization of Mutant/Transgenic Plants by RT-PCR
4.7. RNA-Seq and Data Analysis
4.8. Real-Time Quantitative PCR (qRT PCR)
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ABCB | ATP-binding cassette B protein |
| ARF | auxin response factor |
| AtHB | Arabidopsis thaliana homeobox-leucine zipper protein |
| AtRBK | Arabidopsis thaliana ROP-binding kinase |
| BZR | brassinazole-resistant |
| DEG | differentially expressed gene |
| DELLA | protein domain of the key negative regulators of GA action (DELLA proteins) |
| EIR | ethylene insensitivity of the root |
| FDR | false discovery rate |
| GA | gibberellin |
| ga1 | gibberellic acid 1-gibberellin synthesis mutant |
| GAI | gibberellic acid-insensitive protein |
| GAPC-2 | glyceraldehyde 3-phosphate dehydrogenase C-2 |
| GO | gene ontology |
| GRAS | GA INSENSITIVE (GAI), REPRESSOR of ga1-3 (RGA), and SCARECROW (SCR) protein domain |
| GTP | guanosine-5’-triphosphate |
| HvRBK | Hordeum vulgare ROP-binding kinase |
| LAX | Like auxin1 |
| MtRRK1 | Medicago truncatula ROP-activated Receptor-like Kinase 1 |
| NPF | NRT1/PTR family |
| NRT | nitrate transporter |
| PCR | polymerase chain reaction |
| PIF | phytochrome interacting factor |
| PIN | PINNOID protein |
| PTR | peptide transporter |
| RLCK | receptor-like cytoplasmic kinase |
| RLK | receptor-like kinase |
| ROP | Rho-of-plants |
| SD | short day |
| T-DNA | transfer-DNA |
| TF | transcription factor |
References
- Lehti-Shiu, M.D.; Shiu, S.-H. Diversity, classification and function of the plant protein kinase superfamily. Philos. Trans. R. Soc. B Biol. Sci. 2012, 367, 2619–2639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shiu, S.H.; Bleecker, A.B. Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. USA 2001, 98, 10763–10768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shiu, S.-H.; Karlowski, W.M.; Pan, R.; Tzeng, Y.-H.; Mayer, K.F.X.; Li, W.-H. Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 2004, 16, 1220–1234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, X.; Zhou, J.-M. Receptor-Like Cytoplasmic Kinases: Central players in plant receptor kinase–mediated signaling. Annu. Rev. Plant Biol. 2018, 69, 267–299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jurca, M.E.; Bottka, S.; Fehér, A. Characterization of a family of Arabidopsis receptor-like cytoplasmic kinases (RLCK class VI). Plant Cell Rep. 2008, 27, 739–748. [Google Scholar] [CrossRef]
- Huesmann, C.; Reiner, T.; Hoefle, C.; Preuss, J.; Jurca, M.E.; Domoki, M.; Fehér, A.; Hückelhoven, R. Barley ROP binding kinase1 is involved in microtubule organization and in basal penetration resistance to the barley powdery mildew fungus. Plant Physiol. 2012, 159, 311–320. [Google Scholar] [CrossRef] [Green Version]
- Molendijk, A.J.; Ruperti, B.; Singh, M.K.; Dovzhenko, A.; Ditengou, F.A.; Milia, M.; Westphal, L.; Rosahl, S.; Soellick, T.; Uhrig, J.; et al. A cysteine-rich receptor-like kinase NCRK and a pathogen-induced protein kinase RBK1 are Rop GTPase interactors. Plant J. 2008, 53, 909–923. [Google Scholar] [CrossRef]
- Dorjgotov, D.; Jurca, M.E.; Fodor-Dunai, C.; Szűcs, A.; Ötvös, K.; Klement, É.; Bíró, J.; Fehér, A. Plant Rho-type (Rop) GTPase-dependent activation of receptor-like cytoplasmic kinases in vitro. FEBS Lett. 2009, 583, 1175–1182. [Google Scholar] [CrossRef] [Green Version]
- Reiner, T.; Hoefle, C.; Huesmann, C.; Ménesi, D.; Fehér, A.; Hückelhoven, R. The Arabidopsis ROP-activated receptor-like cytoplasmic kinase RLCK VI_A3 is involved in control of basal resistance to powdery mildew and trichome branching. Plant Cell Rep. 2015, 34, 457–468. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Z.; Manser, E. PAK and other Rho-associated kinases–effectors with surprisingly diverse mechanisms of regulation. Biochem. J. 2005, 386, 201–214. [Google Scholar] [CrossRef]
- Fehér, A.; Lajkó, D.B. Signals fly when kinases meet Rho-of-plants (ROP) small G-proteins. Plant Sci. 2015, 237, 93–107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lajkó, D.B.; Valkai, I.; Domoki, M.; Ménesi, D.; Ferenc, G.; Ayaydin, F.; Fehér, A. In silico identification and experimental validation of amino acid motifs required for the Rho-of-plants GTPase-mediated activation of receptor-like cytoplasmic kinases. Plant Cell Rep. 2018, 37, 627–639. [Google Scholar] [CrossRef] [Green Version]
- Feiguelman, G.; Fu, Y.; Yalovsky, S. ROP GTPases structure-function and signaling pathways. Plant Physiol. 2018, 176, 57–79. [Google Scholar] [CrossRef] [PubMed]
- Enders, T.A.; Frick, E.M.; Strader, L.C. An Arabidopsis kinase cascade influences auxin-responsive cell expansion. Plant J. 2017, 92, 68–81. [Google Scholar] [CrossRef]
- Kleinboelting, N.; Huep, G.; Kloetgen, A.; Viehoever, P.; Weisshaar, B. GABI-Kat SimpleSearch: New features of the Arabidopsis thaliana T-DNA mutant database. Nucleic Acids Res. 2012, 40, D1211–D1215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scholl, R.L.; May, S.T.; Ware, D.H. Seed and molecular resources for Arabidopsis. Plant Physiol. 2000, 124, 1477–1480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rubio, V.; Shen, Y.; Saijo, Y.; Liu, Y.; Gusmaroli, G.; Dinesh-Kumar, S.P.; Deng, X.W. An alternative tandem affinity purification strategy applied to Arabidopsis protein complex isolation. Plant J. 2005, 41, 767–778. [Google Scholar] [CrossRef]
- Zuo, J.; Niu, Q.W.; Chua, N.H. Technical advance: An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 2000, 24, 265–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ivakov, A.; Persson, S. Plant cell shape: Modulators and measurements. Front. Plant Sci. 2013, 4, 439. [Google Scholar] [CrossRef] [Green Version]
- Yamaguchi, S. Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 2008, 59, 225–251. [Google Scholar] [CrossRef]
- Choi, H.; Jeong, S.; Kim, D.S.; Na, H.J.; Ryu, J.S.; Lee, S.S.; Nam, H.G.; Lim, P.O.; Woo, H.R. The homeodomain-leucine zipper ATHB23, a phytochrome B-interacting protein, is important for phytochrome B-mediated red light signaling. Physiol. Plant. 2014, 150, 308–320. [Google Scholar] [CrossRef]
- Hedden, P.; Thomas, S.G. Gibberellin biosynthesis and its regulation. Biochem. J. 2012, 444, 11–25. [Google Scholar] [CrossRef] [Green Version]
- Wulff, N.; Ernst, H.A.; Jørgensen, M.E.; Lambertz, S.; Maierhofer, T.; Belew, Z.M.; Crocoll, C.; Motawia, M.S.; Geiger, D.; Jørgensen, F.S.; et al. An optimized screen reduces the number of GA transporters and provides insights into Nitrate Transporter 1/Peptide Transporter family substrate determinants. Front. Plant Sci. 2019, 10, 1106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiba, Y.; Shimizu, T.; Miyakawa, S.; Kanno, Y.; Koshiba, T.; Kamiya, Y.; Seo, M. Identification of Arabidopsis thaliana NRT1/PTR FAMILY (NPF) proteins capable of transporting plant hormones. J. Plant Res. 2015, 128, 679–686. [Google Scholar] [CrossRef] [PubMed]
- Tal, I.; Zhang, Y.; Jørgensen, M.E.; Pisanty, O.; Barbosa, I.C.R.; Zourelidou, M.; Regnault, T.; Crocoll, C.; Erik Olsen, C.; Weinstain, R.; et al. The Arabidopsis NPF3 protein is a GA transporter. Nat. Commun. 2016, 7, 11486. [Google Scholar] [CrossRef] [PubMed]
- Corratgé-Faillie, C.; Lacombe, B. Substrate (un)specificity of Arabidopsis NRT1/PTR FAMILY (NPF) proteins. J. Exp. Bot. 2017, 68, 3107–3113. [Google Scholar] [CrossRef]
- Kanno, Y.; Oikawa, T.; Chiba, Y.; Ishimaru, Y.; Shimizu, T.; Sano, N.; Koshiba, T.; Kamiya, Y.; Ueda, M.; Seo, M. AtSWEET13 and AtSWEET14 regulate gibberellin-mediated physiological processes. Nat. Commun. 2016, 7, 13245. [Google Scholar] [CrossRef]
- Sun, T.P.; Kamiya, Y. The Arabidopsis GA1 locus encodes the cyclase ent-kaurene synthetase A of gibberellin biosynthesis. Plant Cell 1994, 6, 1509–1518. [Google Scholar]
- Archacki, R.; Buszewicz, D.; Sarnowski, T.J.; Sarnowska, E.; Rolicka, A.T.; Tohge, T.; Fernie, A.R.; Jikumaru, Y.; Kotlinski, M.; Iwanicka-Nowicka, R.; et al. BRAHMA ATPase of the SWI/SNF chromatin remodeling complex acts as a positive regulator of gibberellin-mediated responses in arabidopsis. PLoS ONE 2013, 8, e58588. [Google Scholar] [CrossRef] [Green Version]
- Li, K.; Yu, R.; Fan, L.-M.; Wei, N.; Chen, H.; Deng, X.W. DELLA-mediated PIF degradation contributes to coordination of light and gibberellin signalling in Arabidopsis. Nat. Commun. 2016, 7, 11868. [Google Scholar] [CrossRef] [Green Version]
- Oh, E.; Zhu, J.Y.; Bai, M.Y.; Arenhart, R.A.; Sun, Y.; Wang, Z.Y. Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. eLife 2014, 2014, e03031. [Google Scholar] [CrossRef] [PubMed]
- Vij, S.; Giri, J.; Dansana, P.K.; Kapoor, S.; Tyagi, A.K. The receptor-like cytoplasmic kinase (OsRLCK) gene family in rice: Organization, phylogenetic relationship, and expression during development and stress. Mol. Plant 2008, 1, 732–750. [Google Scholar] [CrossRef]
- Lin, W.; Ma, X.; Shan, L.; He, P. Big Roles of Small Kinases: The Complex Functions of Receptor-like Cytoplasmic Kinases in Plant Immunity and Development. J. Integr. Plant Biol. 2013, 55, 1188–1197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Wu, S.; Liu, Z.; Friml, J. Environmental and endogenous control of cortical microtubule orientation. Trends Cell Biol. 2016, 26, 409–419. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Wang, X.; Sun, J.; Mao, T. Coordinated regulation of hypocotyl cell elongation by light and ethylene through a microtubule destabilizing protein. Plant Physiol. 2018, 176, 678–690. [Google Scholar] [CrossRef] [Green Version]
- Fu, Y.; Xu, T.; Zhu, L.; Wen, M.; Yang, Z. A ROP GTPase signaling pathway controls cortical microtubule ordering and cell expansion in Arabidopsis. Curr. Biol. 2009, 19, 1827–1832. [Google Scholar] [CrossRef] [Green Version]
- Cosgrove, D.J. Loosening of plant cell walls by expansins. Nature 2000, 407, 321–326. [Google Scholar] [CrossRef]
- Van Sandt, V.S.T.; Suslov, D.; Verbelen, J.-P.; Vissenberg, K. Xyloglucan endotransglucosylase activity loosens a plant cell wall. Ann. Bot. 2007, 100, 1467–1473. [Google Scholar] [CrossRef] [Green Version]
- Sadava, D.; Chrispeels, M.J. Hydroxyproline-rich cell wall protein (extensin): Role in the cessation of elongation in excised pea epicotyls. Dev. Biol. 1973, 30, 49–55. [Google Scholar] [CrossRef]
- Passardi, F.; Penel, C.; Dunand, C. Performing the paradoxical: How plant peroxidases modify the cell wall. Trends Plant Sci. 2004, 9, 534–540. [Google Scholar] [CrossRef]
- Bernardo-García, S.; de Lucas, M.; Martínez, C.; Espinosa-Ruiz, A.; Davière, J.-M.; Prat, S. BR-dependent phosphorylation modulates PIF4 transcriptional activity and shapes diurnal hypocotyl growth. Genes Dev. 2014, 28, 1681–1694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Willige, B.C.; Isono, E.; Richter, R.; Zourelidou, M.; Schwechheimer, C. Gibberellin regulates PIN-FORMED abundance and is required for auxin transport–dependent growth and development in Arabidopsis thaliana. Plant Cell 2011, 23, 2184–2195. [Google Scholar] [CrossRef] [Green Version]
- Curtis, M.D.; Grossniklaus, U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003, 133, 462–469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koncz, C.; Martini, N.; Szabados, L.; Hrouda, M.; Bachmair, A.; Schell, J. Specialized vectors for gene tagging and expression studies. In Plant Molecular Biology Manual; Gelvin, S.B., Schilperoort, R.A., Eds.; Springer: Dordrecht, The Netherland, 1994; pp. 53–74. ISBN 978-94-011-0511-8. [Google Scholar]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium -mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [Green Version]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Talbot, M.J.; White, R.G. Cell surface and cell outline imaging in plant tissues using the backscattered electron detector in a variable pressure scanning electron microscope. Plant Methods 2013, 9, 40. [Google Scholar] [CrossRef] [Green Version]
- Urbanová, T.; Tarkowská, D.; Novák, O.; Hedden, P.; Strnad, M. Analysis of gibberellins as free acids by ultra performance liquid chromatography-tandem mass spectrometry. Talanta 2013, 112, 85–94. [Google Scholar] [CrossRef] [Green Version]
- Rittenberg, D.; Foster, G.L. A new procedure for quantitative analysis by isotope dilution, with application to the determination of amino acids and fatty acids. J. Biol. Chem. 1940, 133, 737–744. [Google Scholar]
- Wu, G.; Carville, J.S.; Spalding, E.P. ABCB19-mediated polar auxin transport modulates Arabidopsis hypocotyl elongation and the endoreplication variant of the cell cycle. Plant J. 2016, 85, 209–218. [Google Scholar] [CrossRef] [Green Version]
- Ortuño, A.; Río, J.D.D.; Casas, J.; Serrano, M.; Acosta, M.; Sánchez-Bravo, J. Influence of ACC and Ethephon on cell growth in etiolated lupin hypocotyls. dependence on cell growth state. Biol. Plant. 2008, 33, 81. [Google Scholar] [CrossRef]
- Alabadí, D.; Gil, J.; Blázquez, M.A.; García-Martínez, J.L. Gibberellins repress photomorphogenesis in darkness. Plant Physiol. 2004, 134, 1050–1057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baba, A.I.; Valkai, I.; Labhane, N.M.; Koczka, L.; Andrási, N.; Klement, É.; Darula, Z.; Medzihradszky, K.F.; Szabados, L.; Fehér, A.; et al. CRK5 Protein Kinase Contributes to the Progression of Embryogenesis of Arabidopsis thaliana. Int. J. Mol. Sci. 2019, 20, 6120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, C.-Y.; Krishnakumar, V.; Chan, A.P.; Thibaud-Nissen, F.; Schobel, S.; Town, C.D. Araport11: A complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 2017, 89, 789–804. [Google Scholar] [CrossRef] [PubMed] [Green Version]







| GO Term | Ontology | Description | p-Value | FDR |
|---|---|---|---|---|
| GO:0050896 | P | response to stimulus | 8.6 × 10−66 | 2.7 × 10−63 |
| GO:0006950 | P | response to stress | 1.4 × 10−47 | 2.2 × 10−45 |
| GO:0009605 | P | response to external stimulus | 1.8 × 10−36 | 2.00 × 10−34 |
| GO:0009607 | P | response to biotic stimulus | 1.9 × 10−29 | 1.6 × 10−27 |
| GO:0009628 | P | response to abiotic stimulus | 1.2 × 10−27 | 7.4 × 10−26 |
| GO:0051704 | P | multi-organism process | 2.2 × 10−24 | 1.2 × 10−22 |
| GO:0009719 | P | response to endogenous stimulus | 8.1 × 10−24 | 3.7 × 10−22 |
| GO:0019748 | P | secondary metabolic process | 2.6 × 10−20 | 1.00 × 10−18 |
| GO:0007154 | P | cell communication | 3.7 × 10−15 | 1.3 × 10−13 |
| GO:0009056 | P | catabolic process | 6.3 × 10−12 | 2.00 × 10−10 |
| GO:0007165 | P | signal transduction | 2.8 × 10−11 | 8.3 × 10−10 |
| GO:0009987 | P | cellular process | 2.7 × 10−10 | 7.2 × 10−9 |
| GO:0008152 | P | metabolic process | 2.2 × 10−9 | 5.4 × 10−8 |
| GO:0051179 | P | localization | 3.3 × 10−9 | 7.6 × 10−8 |
| GO:0009991 | P | response to extracellular stimulus | 1.5 × 10−8 | 3.1 × 10−7 |
| GO:0006629 | P | lipid metabolic process | 2.1 × 10−8 | 4.1 × 10−7 |
| GO:0006810 | P | transport | 3.2 × 10−8 | 6.1 × 10−7 |
| GO:0051234 | P | establishment of localization | 4.7 × 10−8 | 8.4 × 10−7 |
| GO:0005975 | P | carbohydrate metabolic process | 2.2 × 10−7 | 3.7 × 10−6 |
| GO:0008219 | P | cell death | 7.9 × 10−6 | 0.00013 |
| GO:0065008 | P | regulation of biological quality | 2.00 × 10−5 | 0.00031 |
| GO:0065007 | P | biological regulation | 2.7 × 10−5 | 0.00039 |
| GO:0042592 | P | homeostatic process | 5.6 × 10−5 | 0.00078 |
| GO:0040007 | P | growth | 0.0009 | 0.012 |
| GO:0009606 | P | tropism | 0.0011 | 0.014 |
| GO:0032502 | P | developmental process | 0.0017 | 0.021 |
| GO:0050789 | P | regulation of biological process | 0.002 | 0.024 |
| GO:0050794 | P | regulation of cellular process | 0.0025 | 0.029 |
| GO:0048856 | P | anatomical structure development | 0.0038 | 0.042 |
| GO:0003824 | F | catalytic activity | 1.3 × 10−25 | 1.3 × 10−23 |
| GO:0005215 | F | transporter activity | 4.2 × 10−9 | 2.1 × 10−7 |
| GO:0016740 | F | transferase activity | 1.9 × 10−8 | 6.4 × 10−7 |
| GO:0008289 | F | lipid binding | 1.2 × 10−7 | 3.00 × 10−6 |
| GO:0030246 | F | carbohydrate binding | 5.7 × 10−6 | 0.00012 |
| GO:0016787 | F | hydrolase activity | 1.2 × 10−5 | 0.0002 |
| GO:0019825 | F | oxygen binding | 1.9 × 10−5 | 0.00027 |
| GO:0016301 | F | kinase activity | 2.5 × 10−5 | 0.00032 |
| GO:0000166 | F | nucleotide binding | 0.00014 | 0.0016 |
| GO:0005488 | F | binding | 0.00021 | 0.0021 |
| GO:0060089 | F | molecular transducer activity | 0.00041 | 0.0034 |
| GO:0004872 | F | receptor activity | 0.00041 | 0.0034 |
| GO:0016772 | F | transferase activity, transferring phosphorus-containing groups | 0.0012 | 0.009 |
| GO:0005576 | C | extracellular region | 8.6 × 10−37 | 1.6 × 10−34 |
| GO:0030312 | C | external encapsulating structure | 6.7 × 10−34 | 4.2 × 10−32 |
| GO:0005618 | C | cell wall | 6.7 × 10−34 | 4.2 × 10−32 |
| GO:0005886 | C | plasma membrane | 7.5 × 10−32 | 3.5 × 10−30 |
| GO:0016020 | C | membrane | 1.3 × 10−25 | 4.9 × 10−24 |
| GO:0005773 | C | vacuole | 4.1 × 10−5 | 0.0013 |
| GO:0005783 | C | endoplasmic reticulum | 8.8 × 10−5 | 0.0023 |
| GO:0012505 | C | endomembrane system | 0.0018 | 0.043 |
| Gene_ID | Annotation | Fold Change | q_Value | Significant |
|---|---|---|---|---|
| DEGs Implicated in GA Metabolism/Response * | ||||
| AT1G26960 | AtHB23 homeobox protein 23 | 2.25 | 0.026242 | yes |
| AT1G74670 | GASA6 Gibberellin-regulated family protein | 2.07 | 0.001941 | yes |
| AT2G37640 | EXP3 Barwin-like endoglucanases superfamily protein | 1.95 | 0.001941 | yes |
| AT5G15230 | GASA4 GAST1 protein homolog 4 | 1.88 | 0.001941 | yes |
| AT2G14900 | AT2G14900 Gibberellin-regulated family protein | 1.70 | 0.001941 | yes |
| AT1G14920 | GAI GRAS family transcription factor family protein | 1.46 | 0.00859 | yes |
| AT3G11280 | AT3G11280 Duplicated homeodomain-like superfamily protein | 0.69 | 0.00859 | yes |
| AT4G19700 | RING SBP (S-ribonuclease binding protein) family protein | 0.68 | 0.038936 | yes |
| AT1G75750 | GASA1 GAST1 protein homolog 1 | 0.56 | 0.003506 | yes |
| AT5G44610 | MAP18 microtubule-associated protein 18 | 0.55 | 0.001941 | yes |
| AT1G02400 | GA2OX6 gibberellin 2-oxidase 6 | 0.58 | 0.044627 | yes |
| DEGs of NPF (NRT1/PTR FAMILY) Gibberellin Transporters ** | ||||
| AT1G52190 | AT1G52190 Major facilitator superfamily protein, NPF1.2 a,b | 1.60 | 0.001941 | yes |
| AT3G16180 | AT3G16180 Major facilitator superfamily protein, NPF 1.1 a | 1.79 | 0.001941 | yes |
| AT5G46050 | PTR3 peptide transporter 3, NPF 5.2 a | 0.55 | 0.001941 | yes |
| AT5G62680 | GTR2 Major facilitator superfamily protein, NPF 2.11 c | 0.65 | 0.031474 | yes |
| DEGs of DELLA Transcription Regulators *** | ||||
| AT1G14920 | GAI GRAS family transcription factor family protein | 1.46 | 0.00859 | yes |
| AT2G01570 | RGL1 RGA-like 1 | 1.25 | 0.483175 | no |
| AT1G66350 | RGA1 GRAS family transcription factor family protein | 1.11 | 0.773976 | no |
| AT3G03450 | RGL2 RGA-like 2 | 1.58 | 0.496428 | no |
| AT5G17490 | RGL3 RGA-like protein 3 | 1.21 | 0.819669 | no |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Valkai, I.; Kénesi, E.; Domonkos, I.; Ayaydin, F.; Tarkowská, D.; Strnad, M.; Faragó, A.; Bodai, L.; Fehér, A. The Arabidopsis RLCK VI_A2 Kinase Controls Seedling and Plant Growth in Parallel with Gibberellin. Int. J. Mol. Sci. 2020, 21, 7266. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21197266
Valkai I, Kénesi E, Domonkos I, Ayaydin F, Tarkowská D, Strnad M, Faragó A, Bodai L, Fehér A. The Arabidopsis RLCK VI_A2 Kinase Controls Seedling and Plant Growth in Parallel with Gibberellin. International Journal of Molecular Sciences. 2020; 21(19):7266. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21197266
Chicago/Turabian StyleValkai, Ildikó, Erzsébet Kénesi, Ildikó Domonkos, Ferhan Ayaydin, Danuše Tarkowská, Miroslav Strnad, Anikó Faragó, László Bodai, and Attila Fehér. 2020. "The Arabidopsis RLCK VI_A2 Kinase Controls Seedling and Plant Growth in Parallel with Gibberellin" International Journal of Molecular Sciences 21, no. 19: 7266. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21197266