Characterization of a Novel Rice Dynamic Narrow-Rolled Leaf Mutant with Deficiencies in Aromatic Amino Acids
Abstract
:1. Introduction
2. Results
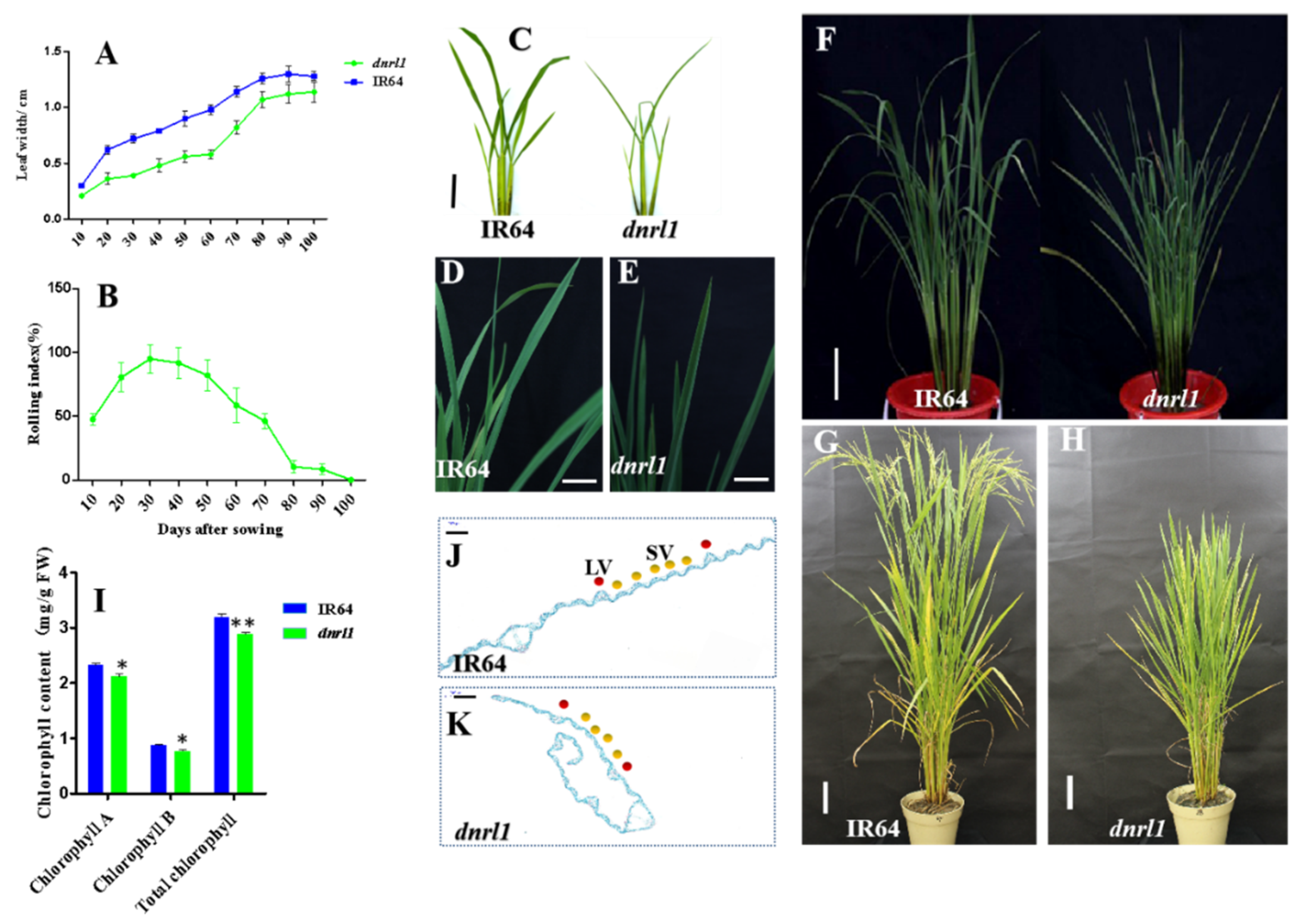
2.1. Phenotype of dnrl1 Mutant
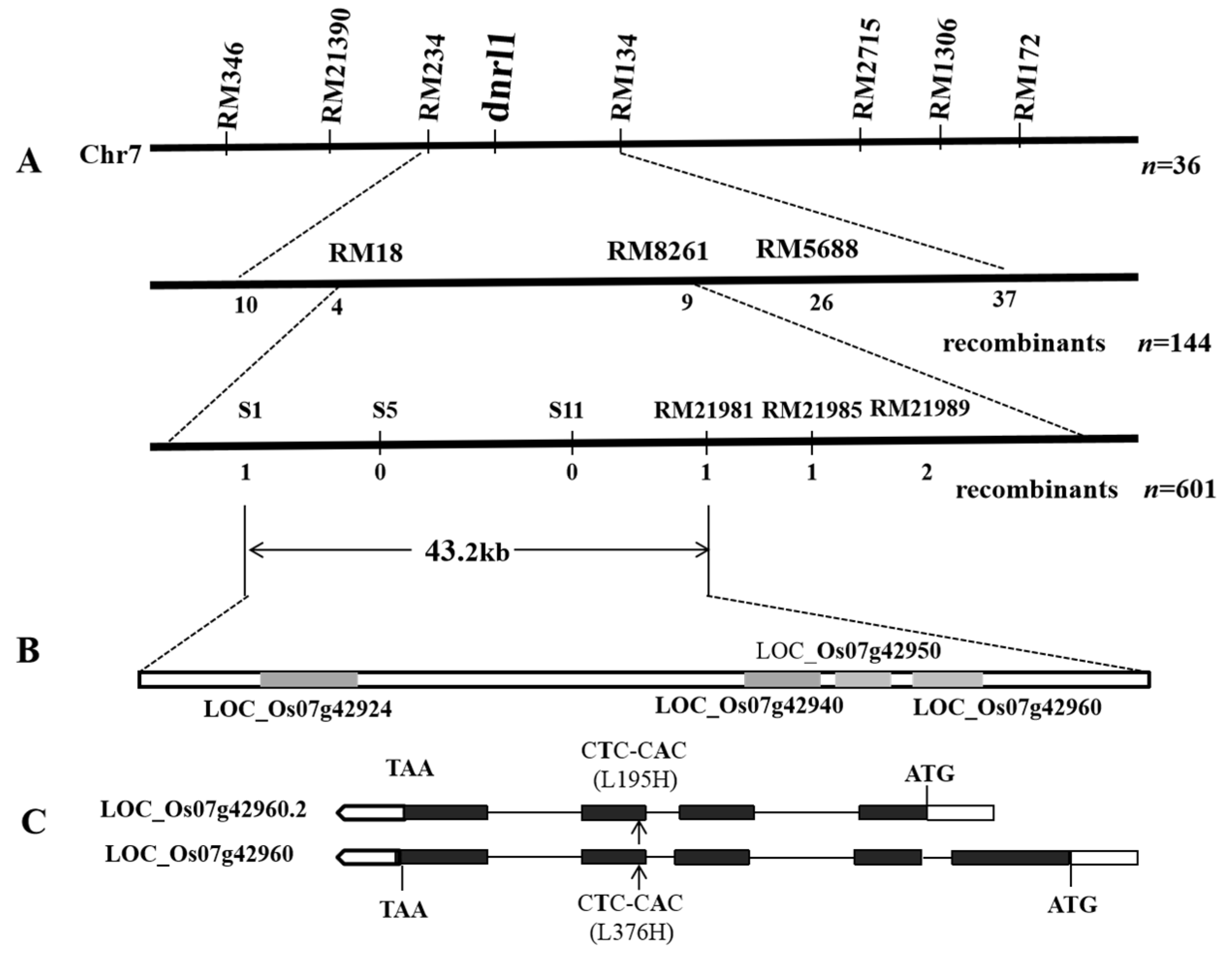
2.2. Genetic Control and Fine Mapping of DNRL1
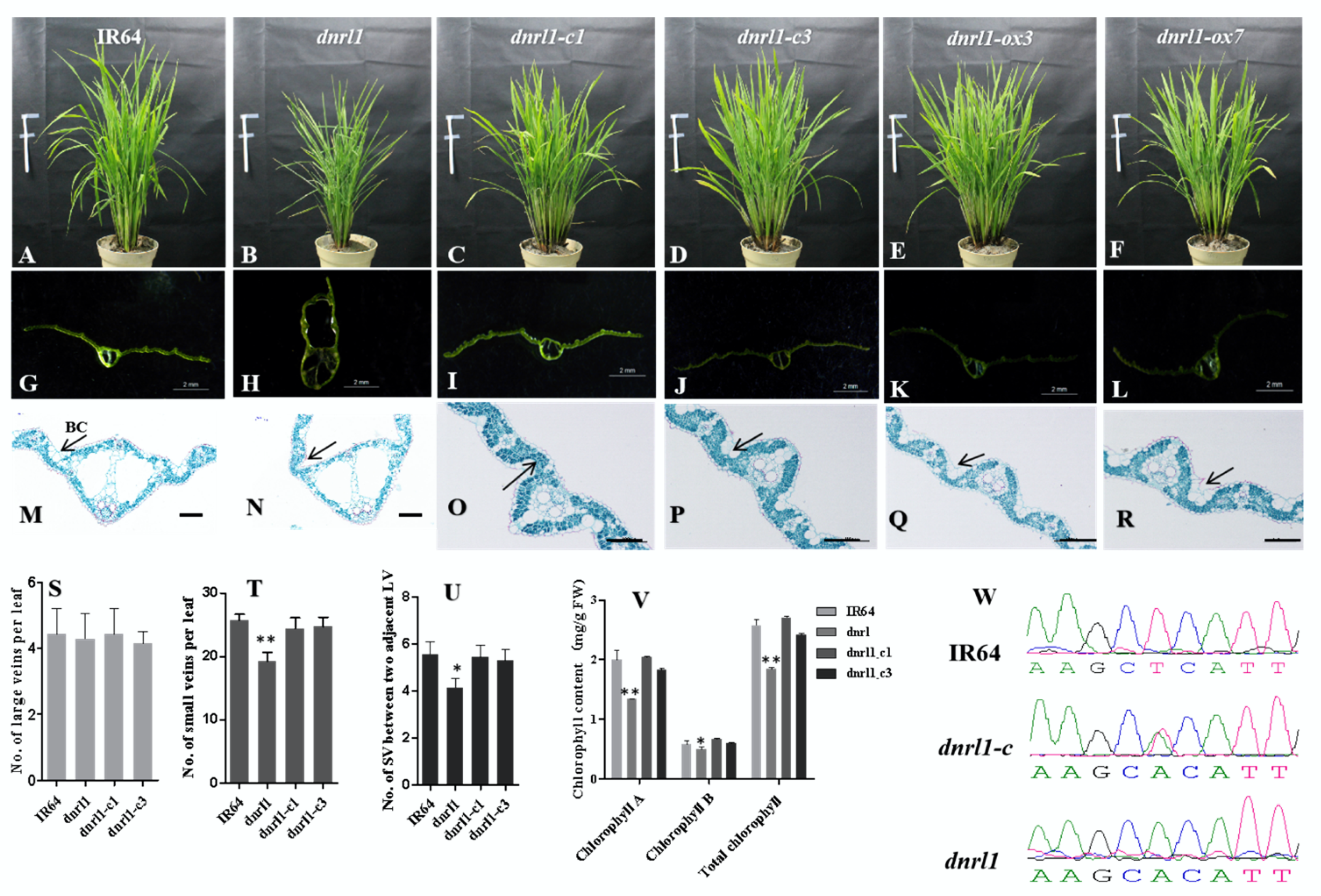
2.3. DNRL1 Complements the dnrl1 Phenotype
2.4. DNRL1 Encodes a DAHP Synth II Enzyme and Modulates Aromatic Amino Acid Synthesis
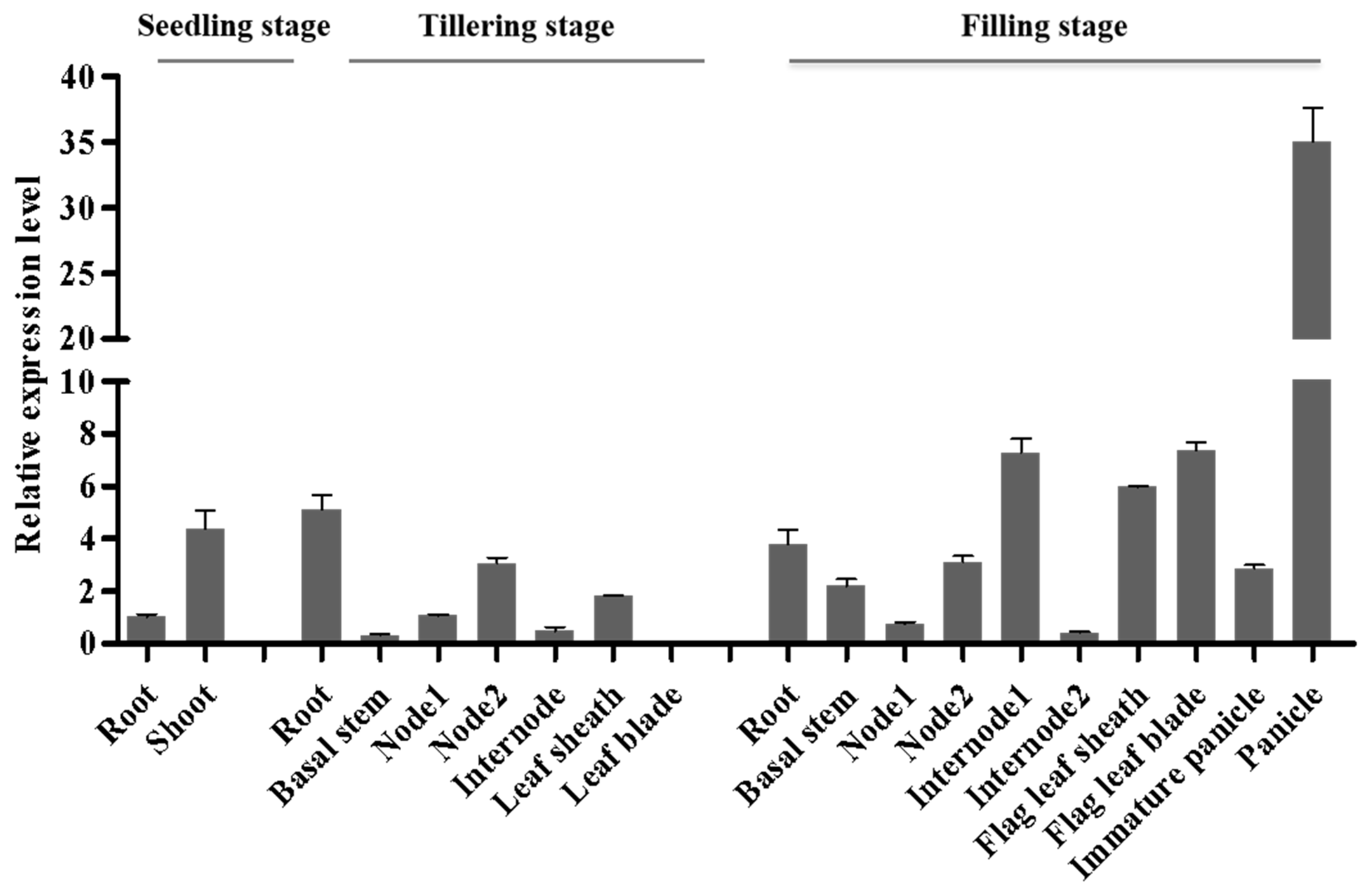
2.5. DNRL1 Expresses Constitutively
2.6. DNRL1-GFP Localizes to Chloroplasts
2.7. Altered Expression of Leaf Morphology and Chlorophyll Biosynthesis-Associated Genes in dnrl1
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Map-Based Cloning of DNRL1
4.3. Genetic Complementation and Over-Expression Analysis
4.4. Measurement of Chlorophyll Content
4.5. Quantitative Reverse Transcription PCR (qRT-PCR)
4.6. Subcellular Localization
4.7. Determination of Amino Acid Content
4.8. Assay of DAHPS Activity
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Tsukaya, H. Mechanism of leaf-shape determination. Annu. Rev. Plant Biol. 2006, 57, 477–496. [Google Scholar] [CrossRef]
- Micol, J. Leaf development: Time to turn over a new leaf? Curr. Opin. Plant Biol. 2009, 12, 9–16. [Google Scholar] [CrossRef]
- Yuan, L. Hybrid rice breeding for super high yield. Hybrid Rice 1997, 12, 1–6, (In Chinese with English Abstract). [Google Scholar]
- Pang, J.; Xie, H.; Qian, M.; Xu, B.; Cai, Y.; Peng, X. Research progress on rice narrow-leaf mutant associated gene. Mol. Plant Breed. 2017, 15, 4879–4887. [Google Scholar]
- Liu, B.; Li, P.; Li, X.; Liu, C.; Cao, S.; Chu, C.; Cao, X. Loss of function of OsDCL1 affects MicroRNA accumulation and causes developmental defects in rice. Plant Physiol. 2005, 139, 296–305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, D.; Liu, M.; Tang, M.; Dong, B.; Wu, D.; Zhang, Z.; Zhou, B. Repression of microrna biogenesis by silencing of OsDCL1 activates the basal resistance to Magnaporthe oryzae in rice. Plant Sci. 2015, 237, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.; Yoo, S.; Zhang, H.; Pandeya, D.; Koh, H.; Hwang, J.; Kim, G.; Paek, N. The rice narrow leaf2 and narrow leaf3 loci encode WUSCHEL-related homeobox 3A (OsWOX3A) and function in leaf, spikelet, tiller and lateral root development. New Phytol. 2013, 198, 1071–1084. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Xu, Q.; Zhu, X.; Qian, Q.; Xue, H. Shallot-like1 is a kanadi transcription factor that modulates rice leaf rolling by regulating leaf abaxial cell development. Plant Cell 2009, 21, 719–735. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.; Fu, Y.; Hu, G.; Si, H.; Cheng, S.; Liu, W. Isolation and characterization of a rice mutant with narrow and rolled leaves. Planta 2010, 232, 313–324. [Google Scholar] [CrossRef]
- Hu, J.; Zhu, L.; Zeng, D.; Gao, Z.; Guo, L.; Fang, Y.; Zhang, G.; Dong, G.; Yan, M.; Liu, J.; et al. Identification and characterization of NARROW AND ROLLED LEAF 1, a novel gene regulating leaf morphology and plant architecture in rice. Plant Mol. Biol. 2010, 73, 283–292. [Google Scholar] [CrossRef]
- Li, M.; Xiong, G.; Li, R.; Cui, J.; Tang, D.; Zhang, B.; Pauly, M.; Cheng, Z.; Zhou, Y. Rice cellulose synthase-like d4 is essential for normal cell-wall biosynthesis and plant growth. Plant J. 2009, 60, 1055–1069. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshikawa, T.; Eiguchi, M.; Hibara, K.; Ito, J.; Nagato, Y. Rice slender leaf 1 gene encodes cellulose synthase-like d4 and is specifically expressed in m-phase cells to regulate cell proliferation. J. Exp. Bot. 2013, 64, 2049–2061. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, Z.; Lin, Z.; Li, Q.; Wu, H.; Xiang, C.; Wang, J. Dnl1, encodes cellulose synthase-like d4, is a major qtl for plant height and leaf width in rice (Oryza sativa L.). Biochem. Biophys. Res. Commun. 2015, 457, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Luan, W.; Liu, Y.; Zhang, F.; Song, Y.; Wang, Z.; Peng, Y.; Sun, Z. OsCD1 encodes a putative member of the cellulose synthase-liked sub-family and is essential for rice plant architecture and growth. Plant Biotechnol. J. 2011, 9, 513–524. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wu, C.; Hu, G.; Xing, L.; Qian, W.; Si, H.; Sun, Z.; Wang, X.; Fu, Y.; Liu, W. Characterization and fine mapping of a novel rice narrow leaf mutant nal9. J. Integr. Plant Biol. 2013, 55, 1016–1025. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Fei, G.; Wu, C.; Wu, F.; Sun, Y.; Chen, M.; Ren, Y.; Zhou, K.; Cheng, Z.; Wang, J.; et al. A rice virescent-yellow leaf mutant reveals new insights into the role and assembly of plastid caseinolytic protease in higher plants. Plant Physiol. 2013, 162, 1867–1880. [Google Scholar] [CrossRef] [Green Version]
- Woo, Y.; Park, H.; Su’udi, M.; Yang, J.; Park, J.; Back, K.; Park, Y.; An, G. Constitutively wilted 1, a member of the rice YUCCA gene family, is required for maintaining water homeostasis and an appropriate root to shoot ratio. Plant Mol. Biol. 2007, 65, 125–136. [Google Scholar] [CrossRef]
- Qi, J.; Qian, Q.; Bu, Q.; Li, S.; Chen, Q.; Sun, J.; Liang, W.; Zhou, Y.; Chu, C.; Li, X.; et al. Mutation of the rice narrow leaf1 gene, which encodes a novel protein, affects vein patterning and polar auxin transport. Plant Physiol. 2008, 147, 1947–1959. [Google Scholar] [CrossRef] [Green Version]
- Jiang, D.; Fang, J.; Lou, L.; Zhao, J.; Yuan, S.; Yin, L.; Sun, W.; Peng, L.; Guo, B.; Li, X. Characterization of a null allelic mutant of the rice nal1 gene reveals its role in regulating cell division. PLoS ONE 2015, 10, e0118169. [Google Scholar] [CrossRef] [Green Version]
- Lin, L.; Zhao, Y.; Liu, F.; Chen, Q.; Qi, J. Narrow leaf 1 (NAL1) regulates leaf shape by affecting cell expansion in rice (Oryza sativa L.). Biochem. Biophys. Res. Commun. 2019, 516, 957–962. [Google Scholar] [CrossRef]
- Fumika, C.; Yukiko, Y.; Toshihiro, K.; Yutaka, S.; Hiro-Yuki, H. Genetic analysis of rice mutants responsible for narrow leaf phenotype and reduced vein number. Genes Genet. Syst. 2016, 91, 235–240. [Google Scholar]
- Fujino, K.; Matsuda, Y.; Ozawa, K.; Nishimura, T.; Koshiba, T.; Fraaije, M.; Sekiguchi, H. Narrow leaf 7 controls leaf shape mediated by auxin in rice. Mol. Genet. Genom. 2008, 279, 499–507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sazuka, T.; Kamiya, N.; Nishimura, T.; Ohmae, K.; Sato, Y.; Imamura1, K.; Nagato, Y.; Koshiba, T.; Nagamura, Y.; Ashikari1, M.; et al. A rice tryptophan deficient dwarf mutant, tdd1, contains a reduced level of indole acetic acid and develops abnormal flowers and organless embryos. Plant J. 2009, 60, 227–241. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wu, S.; Jiang, L.; Wang, J.; Zhang, X.; Guo, X.; Wu, C.; Wan, J. A detailed analysis of the leaf rolling mutant sll2 reveals complex nature in regulation of bulliform cell development in rice (Oryza sativa L.). Plant Biol. 2015, 17, 437–448. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Zhao, L.; Liu, F.; Wu, Y.; Zhu, Z.; Sun, C.; Tan, L. Narrow and rolled leaf 2 regulates leaf shape, male fertility, and seed size in rice. J. Integr. Plant Biol. 2016, 58, 983–996. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, M.; Liu, K.; Tang, D.; Sun, M.; Li, Y.; Shen, Y.; Du, G.; Cheng, Z. Semi-rolled leaf2 modulates rice leaf rolling by regulating abaxial side cell differentiation. J. Exp. Bot. 2016, 67, 2139–2150. [Google Scholar] [CrossRef] [Green Version]
- Ma, L.; Sang, X.; Zhang, T.; Yu, Z.; Li, Y.; Zhao, F.; Wang, Z.; Wang, Y.; Yu, P.; Wang, N.; et al. Abnormal vascular bundles regulates cell proliferation and procambium cell establishment during aerial organ development in rice. New Phytol. 2017, 213, 275–286. [Google Scholar] [CrossRef] [Green Version]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.; Lu, S.; Chitsaz, F.; Derbyshire, M.; Geer, R.; Gonzales, N.; et al. Cdd/sparcle: Functional classification of proteins via subfamily domain architectures. Nucl. Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef]
- Sato, K.; Mase, K.; Nakano, Y.; Nishikubo, N.; Sugita, R.; Tsuboi, Y.; Kajita, S.; Zhou, J.; Kitano, H.; Katayama, Y. 3-Deoxy-d-arabino-heptulosonate 7-phosphate synthase is regulated for the accumulation of polysaccharide-linked hydroxycinnamoyl esters in rice (Oryza sativa L.) internode cell walls. Plant Cell Rep. 2006, 25, 676–688. [Google Scholar] [CrossRef]
- Chen, W.; Sheng, Z.; Cai, Y.; Li, Q.; Wei, X.; Xie, L.; Jiao, G.; Shao, G.; Tang, S.; Wang, J.; et al. Rice morphogenesis and chlorophyll accumulation is regulated by the protein encoded by NRL3 and its interaction with NAL9. Front. Plant Sci. 2019, 10, 175. [Google Scholar] [CrossRef]
- Xu, J.; Wang, L.; Qian, Q.; Zhang, G. Research advance in molecule regulation mechanism of leaf morphogenesis in Rice (Oryza sativa L.). Acta Agron. Sin. 2013, 39, 767–774, (In Chinese with English Abstract). [Google Scholar] [CrossRef]
- Tohge, T.; Watanabe, M.; Hoefgen, R.; Fernie, A. Shikimate and phenylalanine biosynthesis in the green lineage. Front. Plant Sci. 2013, 4, 62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corea, O.; Ki, C.; Cardenas, C.; Kim, S.; Brewer, S.; Patten, A.; Davin, L.; Lewis, N. Arogenate dehydratase isoenzymes profoundly and differentially modulate carbon flux into lignins. J. Biol. Chem. 2012, 287, 11446–11459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maeda, H.; Dudareva, N. The shikimate pathway and aromatic amino acid biosynthesis in plants. Annu. Rev. Plant Biol. 2012, 63, 73–105. [Google Scholar] [CrossRef] [PubMed]
- Yasui, Y.; Ohmori, Y.; Takebayashi, Y.; Sakakibara, H.; Hirano, H. WUSCHEL-RELATED HOMEOBOX4 acts as a key regulator in early leaf development in rice. PLoS Genet. 2018, 14, e1007365. [Google Scholar] [CrossRef]
- He, P.; Wang, X.; Zhang, X.; Jiang, Y.; Tian, W.; Zhang, X.; Li, Y.; Sun, Y.; Xie, J.; Ni, J.; et al. Short and narrow flag leaf1, a GATA zinc finger domain-containing protein, regulates flag leaf size in rice (Oryza sativa). BMC Plant Biol. 2018, 18, 273. [Google Scholar] [CrossRef]
- Wu, J.; Wu, C.; Lei, C.; Baraoidan, M.; Bordeos, A.; Madamba, M.; Ramos-Pamplona, M.; Mauleon, R.; Portugal, A.; Ulat, V.; et al. Chemical and irradiation induced mutants of indica rice IR64 for forward and reverse genetics. Plant Mol. Biol. 2005, 59, 85–97. [Google Scholar] [CrossRef]
- Lu, Y.; Zheng, K. A simple method for isolation of rice DNA. Chin. J. Rice Sci. 1992, 6, 47–48. [Google Scholar]
- Shi, Y.; Chen, J.; Liu, W.; Huang, Q.; Shen, B.; Leung, H.; Wu, J. Genetic analysis and gene mapping of a new rolled-leaf mutant in rice (Oryza sativa L.). Sci. China Ser. C Life Sci. 2009, 52, 885–890. [Google Scholar] [CrossRef]
- Hiei, Y.; Komari, T. Agrobacterium-mediated transformation of rice using immature embryos or calli induced from mature seed. Nat. Protoc. 2008, 3, 824–834. [Google Scholar] [CrossRef]
- Arnon, D. Copper enzymes in isolated chloroplasts polyphenoloxidase in beta vulgaris. Plant Physiol. 1949, 24, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmittgen, T.; Livak, K. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Su, J.; Duan, S.; Ao, Y.; Dai, J.; Liu, J.; Wang, P.; Li, Y.; Liu, B.; Feng, D.; et al. A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 2011, 7, 30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mei, X.; Chen, Y.; Zhang, L.; Fu, X.; Wei, Q.; Grierson, D.; Zhou, Y.; Huang, Y.; Dong, F.; Yang, Z. Dual mechanisms regulating glutamate decarboxylases and accumulation of gamma-aminobutyric acid in tea (Camellia sinensis) leaves exposed to multiple stresses. Sci. Rep. 2016, 6, 23685. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Fu, X.; Mei, X.; Zhou, Y.; Cheng, S.; Zeng, L.; Dong, F.; Yang, Z. Proteolysis of chloroplast proteins is responsible for accumulation of free amino acids in dark-treated tea (Camellia sinensis) leaves. J. Proteom. 2017, 157, 10–17. [Google Scholar] [CrossRef]
- Wang, Y.; Zhu, L.; Li, J. A new method for enzyme assay of the activity of 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase. J. Southwest Agri. Univ. (Nat. Sci.) 2006, 28, 40–44. [Google Scholar]



| Cross | F1 Phenotype | F2 | P(3:1) | χ2 0.05 | ||
|---|---|---|---|---|---|---|
| Total No. of Plants | No. of Flat Leaf Plants | No. of Narrow Rolled-Leaf Plants | ||||
| dnrll/IR64 | Flat leaf | 833 | 632 | 201 | 0.56 | 0.34 |
| dnrll/Moroberekan | Flat leaf | 2353 | 1752 | 601 | 0.54 | 0.37 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.; Shi, Y.; Zhang, X.; Xu, X.; Wu, J.-L. Characterization of a Novel Rice Dynamic Narrow-Rolled Leaf Mutant with Deficiencies in Aromatic Amino Acids. Int. J. Mol. Sci. 2020, 21, 1521. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21041521
Wang H, Shi Y, Zhang X, Xu X, Wu J-L. Characterization of a Novel Rice Dynamic Narrow-Rolled Leaf Mutant with Deficiencies in Aromatic Amino Acids. International Journal of Molecular Sciences. 2020; 21(4):1521. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21041521
Chicago/Turabian StyleWang, Huimei, Yongfeng Shi, Xiaobo Zhang, Xia Xu, and Jian-Li Wu. 2020. "Characterization of a Novel Rice Dynamic Narrow-Rolled Leaf Mutant with Deficiencies in Aromatic Amino Acids" International Journal of Molecular Sciences 21, no. 4: 1521. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21041521





